From Biomarker to Beacon: Designing and Deploying Nanonetwork Alarm Systems for Early Disease Detection
This article provides a comprehensive exploration of alarm-system nanonetworks engineered for biomarker detection.
From Biomarker to Beacon: Designing and Deploying Nanonetwork Alarm Systems for Early Disease Detection
Abstract
This article provides a comprehensive exploration of alarm-system nanonetworks engineered for biomarker detection. Targeting researchers, scientists, and drug development professionals, we detail the foundational concepts of these bio-inspired communication systems, including their core components like biosensors, nano-transceivers, and receivers. We delve into methodological blueprints for network design, signal processing, and *in vitro*/*in vivo* applications. Critical challenges such as signal interference, biocompatibility, and power constraints are addressed with practical troubleshooting and optimization strategies. Finally, we present a rigorous framework for validating network performance, comparing technological platforms (e.g., DNA-based vs. synthetic nanoparticle networks), and assessing their clinical translatability. This guide synthesizes current research to advance the development of precise, proactive diagnostic tools.
Decoding the Blueprint: Core Components and Principles of Biomarker Alarm Nanonetworks
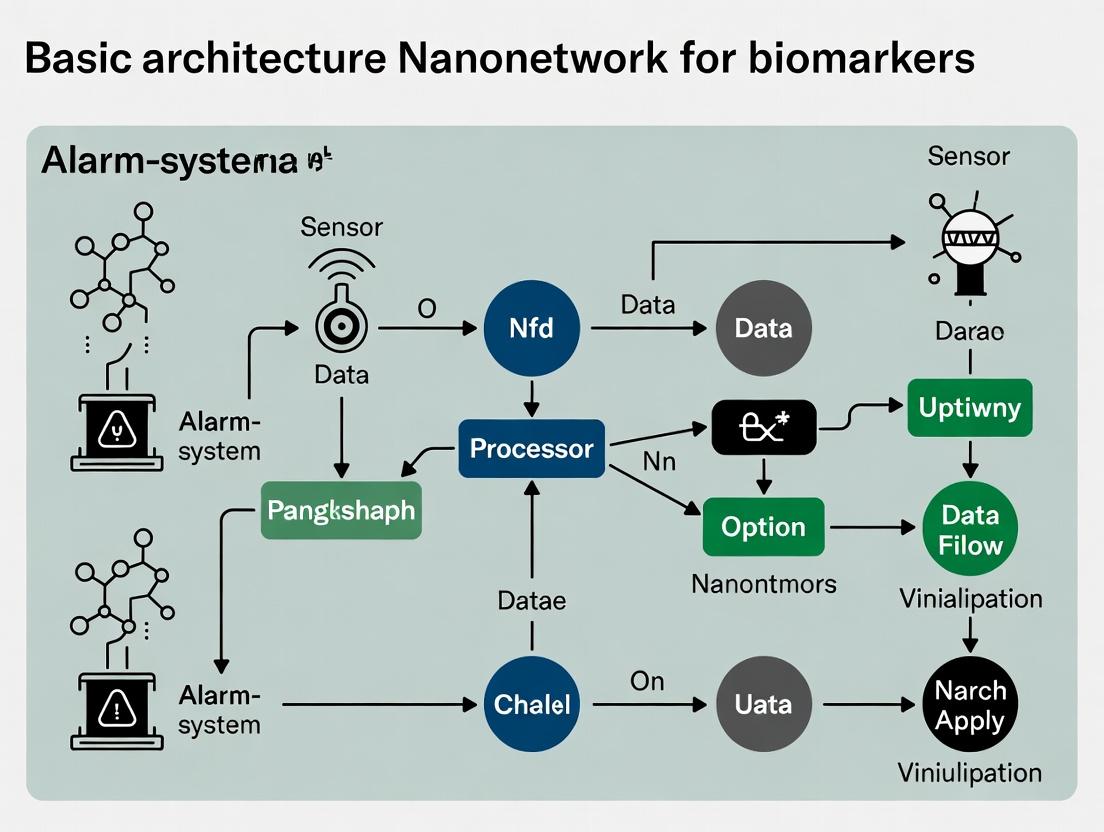
A Biomarker Alarm-System Nanonetwork is an integrated, engineered system comprising nanoscale components (synthetic or bio-hybrid) designed for continuous, in vivo monitoring of disease-specific molecular biomarkers. Upon detection of a pathological concentration threshold, the network autonomously triggers a multi-stage, amplified signal—an "alarm"—communicatable to external devices or capable of initiating a therapeutic response. This whitepaper details its core architecture and operational principles within the broader thesis of foundational research for such systems.
Core Architectural Framework
The basic architecture is a hierarchical network with distinct functional layers, enabling sensing, computation, communication, and actuation.
Table 1: Core Functional Layers of the Alarm-System Nanonetwork
| Layer | Primary Function | Key Nanoscale Components | Output |
|---|---|---|---|
| Sensing | Target biomarker recognition and binding. | Functionalized nanoparticles, engineered nanopores, DNA/RNA aptamers, molecular imprinting polymers. | Biomarker-binding event transduced into a chemical or conformational change. |
| Signal Transduction & Amplification | Convert binding event into a scalable, propagatable signal. | Enzyme cascades (e.g., horseradish peroxidase), nanoparticle quenching/de-quenching, DNAzyme networks, biocatalytic circuits. | Amplified chemical output (e.g., fluorescence, chemiluminescence, ionic flux). |
| Communication & Networking | Relay signal between nodes and to an external interface. | Diffusive molecular communication (calcium waves, ROS species), Förster resonance energy transfer (FRET) chains, wireless electromagnetic (nanoscale antenna). | Coordinated network response surpassing single-node detection limits. |
| Actuation & Reporting | Generate a readable alarm or primary therapeutic effect. | Release of reporter molecules (dyes, peptides), generation of gas bubbles (for ultrasound), triggered drug release from nanocarriers. | Externally detectable signal (e.g., colorimetric urine change, MRI contrast) or localized pharmacological action. |
Key Experimental Protocols
Protocol 1: In Vitro Validation of a Protease-Activated FRET Nanosensor Network
- Objective: To demonstrate network-based signal amplification for a disease-specific protease biomarker (e.g., MMP-9).
- Materials: See "The Scientist's Toolkit" below.
- Methodology:
- Nanosensor Synthesis: Conjugate donor (Cy3) and acceptor (Cy5) fluorophores to a peptide substrate linker specific for MMP-9 cleavage. Attach this construct to a 20nm PEGylated quantum dot (QD) core.
- Network Formation: Mix QD-sensors with cationic liposomes to facilitate aggregation into a nanonetwork cluster via electrostatic interaction. Characterize cluster size using dynamic light scattering (DLS).
- Signal Acquisition: Incubate the nanonetwork with recombinant MMP-9 (10 nM-1 µM range) in a physiological buffer (pH 7.4) at 37°C.
- Data Measurement: Use a fluorescence plate reader to monitor time-dependent loss of FRET (increase in donor emission at 570nm, decrease in acceptor emission at 670nm upon excitation at 530nm). Compare signal kinetics and amplitude against dispersed, non-networked sensors.
Protocol 2: Evaluation of a Glucose-Responsive DNAzyme Cascade for Alarm Triggering
- Objective: To implement a nucleic acid-based amplification circuit that triggers a colorimetric alarm upon sensing hyperglycemia.
- Materials: DNAzyme sequences (8-17E), glucose oxidase (GOx), hemin, horseradish peroxidase (HRP), ABTS substrate, synthetic urine matrix.
- Methodology:
- Circuit Assembly: Mix the substrate strand for the DNAzyme with the enzyme strand in a buffer containing hemin to form the active G-quadruplex DNAzyme structure.
- Integration with Biomarker Sensor: Incorporate GOx into the system. GOx converts glucose to gluconic acid and H₂O₂.
- Cascade Activation: The generated H₂O₂ acts as the co-substrate for the HRP-mimicking DNAzyme.
- Alarm Readout: Add ABTS (2,2'-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)). Oxidation by the DNAzyme-H₂O₂ system produces a green-colored product, measurable at 405-420 nm. Calibrate against glucose concentrations (5-30 mM).
Visualizing Signaling Pathways & Workflows
(Diagram Title: Core Alarm Nanonetwork Signal Pathway)
(Diagram Title: Biomarker Alarm System Development Workflow)
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Prototype Development
| Item | Function in Research | Example & Rationale |
|---|---|---|
| Functionalized Nanoparticles | Core sensing platform. | Gold Nanorods (AuNRs): High surface-area-to-volume ratio for biomarker capture; tunable plasmonic properties for photothermal signal transduction. |
| DNA Aptamers / DNAzymes | High-specificity recognition and catalytic elements. | SELEX-derived Aptamer for PSA: Provides synthetic, stable alternative to antibodies for prostate-specific antigen detection in sensor design. |
| Fluorescent Reporters (FRET Pairs) | For optical signal generation and intra-network communication. | Cy3-Cy5 FRET Pair: Attached via cleavable peptide linker; cleavage by target protease disrupts FRET, generating an optical alarm signal. |
| Enzyme Cascades | Provides intrinsic biochemical signal amplification. | Glucose Oxidase (GOx) + Horseradish Peroxidase (HRP): GOx produces H₂O₂ from glucose; HRP uses H₂O₂ to oxidize a substrate, creating a colorimetric/chemiluminescent readout. |
| Synthetic Biological Matrices | For testing in physiologically relevant conditions. | Artificial Interstitial Fluid / Urine: Validates sensor performance against complex backgrounds with ions, proteins, and pH variations, prior to in vivo studies. |
| Animal Disease Models | For ultimate in vivo validation of alarm function. | Transgenic Mouse Model of Colitis (e.g., IL-10 knockout): Provides a living system to test nanonetworks for biomarkers like TNF-α or calprotectin in real-time. |
This whitepaper details the foundational architecture for an alarm-system nanonetwork designed for biomarker research. The system's core objective is the real-time, in situ detection of specific molecular biomarkers, triggering a coordinated, amplifiable signal to an external receiver. This architecture is critical for advancing drug development, enabling researchers to monitor therapeutic efficacy and disease progression at the molecular level within model organisms or in vitro systems.
Architectural Breakdown of the Core Triad
Biosensors: The Molecular Recognition Element
Biosensors are the network's frontline, comprising engineered biological or synthetic components that bind a target biomarker with high specificity.
Key Design Principles:
- Target: Proteins (e.g., enzymes, cytokines), nucleic acids (miRNA, mRNA), or small molecules.
- Transduction Mechanism: Binding induces a conformational change, cleavage event, or release of a reporter molecule.
- Common Formats: Aptamers, engineered proteins (antibody fragments, DARPins), or allosteric ribozymes.
Nano-Nodes: The Signal Processor and Transmitter
Nano-nodes are nanoscale devices (often synthetic or hybrid particles) that interface with biosensors. They convert the molecular binding event into a transmissible signal.
Primary Functions:
- Signal Amplification: Catalytically generate many secondary messenger molecules per binding event.
- Signal Encoding: Modulate the signal's identity (e.g., type of molecule, pulse frequency) based on biomarker concentration or identity.
- Local Communication: Diffuse or actively relay signals to nearby nano-nodes or the Hub.
Hub/Receiver: The Signal Aggregator and External Interface
The Hub is a centralized nano-device or modified cell that collects signals from multiple nano-nodes. The Receiver is the macroscale instrument that detects the Hub's output.
Hub Operations:
- Signal Integration: Summarizes inputs from a network region.
- Noise Filtering: Applies thresholds to reduce false positives.
- Macro-Signal Generation: Produces an externally detectable signal (e.g., magnetic, acoustic, radiofrequency, or strong fluorescent pulse).
Table 1: Performance Metrics of Current Nanonetwork Components (Representative Data)
| Component | Metric | Typical Range (Current Systems) | Target for Alarm Systems |
|---|---|---|---|
| Biosensor | Dissociation Constant (Kd) | pM - nM | < 1 nM |
| Response Time | Seconds - Minutes | < 60 Seconds | |
| Specificity (Cross-Reactivity) | 5-15% | < 1% | |
| Nano-Node | Signal Amplification Factor | 10^2 - 10^4 per event | > 10^5 per event |
| Communication Range | 1 - 20 μm | 50 - 100 μm | |
| Power Source (if synthetic) | Biochemical / External (e.g., magnetic, ultrasonic) | Endogenous biochemical | |
| Hub/Receiver | Signal-to-Noise Ratio (SNR) | 10 - 30 dB | > 40 dB |
| Detection Limit (Biomarker Conc.) | nM - pM in vitro | fM in complex media | |
| Latency (Event to Readout) | Minutes - Hours | < 10 Minutes |
Table 2: Comparison of Primary Signaling Modalities for Nanonetworks
| Modality | Example Messenger | Advantages | Disadvantages for In Vivo Use |
|---|---|---|---|
| Molecular Diffusion | Calcium ions, IP3, DNA strands | Biocompatible, no external power needed. | Slow, subject to enzymatic degradation. |
| Acoustic | Pressure waves | Good tissue penetration, tunable frequency. | Low spatial resolution, requires external transducer. |
| Magnetic | Superparamagnetic nanoparticle (SPION) rotation | Deep tissue penetration, low background noise. | Requires strong external magnetic field generators. |
| Optical | FRET, Bioluminescence | High spatiotemporal resolution, multiplexing. | Limited tissue penetration, autofluorescence. |
| Radiofrequency/EM | Engineered nanoparticle resonance | Potential for deep penetration. | Technical challenges in miniaturization and control. |
Detailed Experimental Protocols
Protocol 4.1:In VitroValidation of a Protease-Activated Biosensor-Nano-Node Assembly
Objective: To test the activation and amplification kinetics of a nano-node triggered by MMP-9 protease cleavage.
Materials: See "Scientist's Toolkit" (Section 6). Procedure:
- Functionalization: Immobilize the quenched fluorescent peptide substrate (biosensor) onto the surface of the liposomal nano-node (pre-loaded with Texas Red dextran) via a streptavidin-biotin linkage.
- Baseline Measurement: Aliquot the functionalized nano-nodes into a 96-well plate. Acquire baseline fluorescence (Ex/Em: 488/520 nm for FRET quench; 595/615 nm for Texas Red) using a plate reader.
- Stimulation: Add recombinant human MMP-9 to experimental wells at final concentrations ranging from 0.1 nM to 100 nM. Use buffer-only controls.
- Kinetic Readout: Immediately initiate kinetic fluorescence measurements every 30 seconds for 2 hours at 37°C.
- Data Analysis: Plot fluorescence intensity over time. Calculate the rate of signal increase (slope) for each MMP-9 concentration. Determine the limit of detection (LOD) and the effective amplification factor (ratio of Texas Red signal increase to cleaved peptide molecules).
Protocol 4.2: Testing Hub-Based Signal Integration in a Microfluidic Chamber
Objective: To demonstrate that a hub particle can sum inputs from multiple, spatially separated nano-nodes.
Materials: Streptavidin-coated magnetic hub particles, two populations of nano-nodes (emitting distinct DNA "Z" and "Y" strands upon activation), microfluidic mixing device, qPCR system. Procedure:
- Compartmentalized Activation: Load the microfluidic device. In one channel, introduce nano-node population "Z" with its target biomarker. In a separate, parallel channel, introduce nano-node population "Y" with a different target. Allow activation to proceed for 15 minutes.
- Hub Introduction & Capture: Mix the effluents from both channels in a central chamber containing the hub particles. The hub surface is functionalized with complementary DNA strands to capture "Z" and "Y" output strands.
- Incubation & Washing: Incubate for 30 minutes to allow hybridization. Apply a magnetic field to sequester hub particles and wash away unbound strands.
- Hub Interrogation: Elute the bound DNA strands from the hub particles. Quantify the concentrations of "Z" and "Y" strands via qPCR using unique primer sets.
- Analysis: Correlate the ratio and quantity of "Z" and "Y" strands on the hub to the original biomarker concentrations in the separate channels, demonstrating integrative capture.
Visualizations (Graphviz Diagrams)
Diagram 1: Core Triad Signaling Pathway Flow
Diagram 2: General Experimental Workflow for Validation
The Scientist's Toolkit: Key Research Reagent Solutions
| Item / Reagent | Function in Nanonetwork Research | Example Product / Material |
|---|---|---|
| Functionalized Liposomes / Polymersomes | Serve as versatile nano-node cores for encapsulating reporters and surface-presenting biosensors. | DSPC/Cholesterol liposomes, PEG-PLGA polymersomes. |
| Streptavidin-Biotin Conjugation System | Provides a robust, high-affinity link for attaching biosensors (e.g., biotinylated aptamers) to nano-node surfaces. | Streptavidin-coated magnetic beads, EZ-Link Sulfo-NHS-Biotin. |
| FRET-Based Peptide Substrates | Act as cleavable biosensors for protease biomarkers; cleavage disrupts FRET, generating a signal. | Custom peptides with donor/acceptor pairs (e.g., FAM/QXL). |
| DNA Oligonucleotide "Toehold" Switches | Used as programmable, amplifiable biosensors and for inter-node communication via strand displacement. | Custom ssDNA from IDT or Sigma. |
| Recombinant Target Biomarkers | Essential positive controls for calibrating and validating biosensor response in vitro. | Recombinant proteins (e.g., R&D Systems, Abcam). |
| Quantum Dots (QDs) / Upconversion NPs | Act as stable, bright optical reporters within nano-nodes or hubs for deep-tissue signal potential. | CdSe/ZnS QDs, NaYF4:Yb,Er UCNPs. |
| Microfluidic Mixing/Encapsulation Device | Enables precise assembly of nano-network components and testing under controlled flow conditions. | Dolomite Microfluidic Chips. |
| Plate Reader with Kinetic Capability | For high-throughput, time-resolved measurement of optical signals (fluorescence, luminescence). | BioTek Synergy H1, BMG CLARIOstar. |
This whitepaper details the fundamental mechanisms by which the binding of a target biomarker initiates a sequence of nanoscale events, culminating in a detectable signal within an alarm-system nanonetwork. This process is the cornerstone of next-generation diagnostic and drug development platforms.
Within the proposed architecture for an alarm-system nanonetwork, individual nodes are engineered nanostructures designed for specific biomarker surveillance. The "alarm" is a cascade of signal translation events, transforming molecular recognition into a transmissible output. This document deconstructs the core cascade following biomarker binding.
The Core Signaling Cascade: A Stepwise Deconstruction
The cascade follows a generalizable pathway from recognition to signal generation.
Stage 1: Biomarker Recognition and Conformational Change
The initial binding event occurs at a biorecognition element (e.g., an antibody, aptamer, or molecularly imprinted polymer). Binding energy induces a precise conformational rearrangement in the receptor or the surrounding nanostructure.
Stage 2: Proximal Transducer Activation
The conformational change alters the local chemical or physical environment, activating a proximal transducer. This can involve:
- Enzymatic Activation: An enzyme co-localized with the receptor becomes active or accesses its substrate.
- Energy Transfer Modulation: The distance or orientation between a donor and acceptor (e.g., FRET pair) changes, altering energy transfer efficiency.
- Electron Transfer Gate: Binding opens or closes a pathway for electron flow in an electrochemical sensor.
- Steric Shield Removal: Binding displaces a quenching molecule or unveils a reactive site.
Stage 3: Signal Amplification
To detect low-abundance biomarkers, the activated transducer triggers an amplification loop.
- Catalytic Amplification: A single activated enzyme generates thousands of product molecules (e.g., horseradish peroxidase with chromogenic substrate).
- Nanoparticle Growth: The initial product catalyzes the deposition of metal onto a nanoparticle seed, dramatically increasing its size and optical cross-section.
- Cascade Reaction: The product of the first reaction is a catalyst or cofactor for a second, orthogonal reaction.
Stage 4: Output Signal Generation
The amplified intermediate is converted into a final, transmissible signal within the nanonetwork.
- Optical: Emission of light at a specific wavelength (fluorescence, chemiluminescence) or a shift in plasmonic resonance (color change).
- Magnetic: Aggregation of superparamagnetic nanoparticles alters T2 relaxation time for MRI detection.
- Electrochemical: Generation or consumption of redox-active molecules produces a measurable current.
- Acoustic: Changes in nanoparticle mass or stiffness upon binding affect ultrasound backscatter.
Table 1: Quantitative Parameters of Common Signal Translation Modalities
| Modality | Typical Biomarker Kd (M) | Amplification Factor | Limit of Detection (Molar) | Time to Signal (min) |
|---|---|---|---|---|
| Catalytic Colorimetric | 10⁻⁹ – 10⁻¹² | 10³ – 10⁶ | 10⁻¹² – 10⁻¹⁵ | 5 – 30 |
| Fluorescence (Direct) | 10⁻⁹ – 10⁻¹² | 1 – 10 | 10⁻¹⁰ – 10⁻¹² | < 1 |
| Fluorescence (FRET) | 10⁻⁹ – 10⁻¹² | 1 – 10 | 10⁻¹¹ – 10⁻¹³ | 1 – 5 |
| Electrochemical (Amperometric) | 10⁻⁹ – 10⁻¹² | 10² – 10⁵ | 10⁻¹² – 10⁻¹⁵ | 2 – 15 |
| Plasmonic Shift (LSPR) | 10⁻⁹ – 10⁻¹² | 1 – 10² | 10⁻¹² – 10⁻¹⁵ | 1 – 10 |
Experimental Protocol: Validating an Aptamer-Based FRET Cascade
This protocol outlines a method to validate a conformational-change-driven cascade using a dye-quencher labeled aptamer.
Objective: To demonstrate that target biomarker binding induces a conformational shift, separating a fluorophore from a quencher, resulting in a measurable fluorescence increase.
Materials: See The Scientist's Toolkit below.
Procedure:
- Aptamer Conjugation: Reconstitute the thiol-modified DNA aptamer in PBS buffer (pH 7.4). Reduce disulfide bonds using 10 mM TCEP for 1 hour. Purify via desalting column.
- Dye/Quencher Labeling: React the reduced aptamer with a 10:1 molar excess of maleimide-functionalized fluorophore (Cy3) for 2 hours in the dark. Purify via HPLC to isolate single-labeled strands. Hybridize the complementary quencher strand (with Iowa Black RQ) at a 1.2:1 ratio in annealing buffer by heating to 95°C for 5 min and slowly cooling to 25°C over 45 min.
- Sensor Characterization: Dilute the dual-labeled aptamer construct to 100 nM in assay buffer. Acquire a fluorescence emission scan (excitation 550 nm, emission 560-750 nm) to establish the baseline quenched signal.
- Target Binding Assay: Aliquot 100 µL of the sensor solution into a 96-well plate. Add 10 µL of serial dilutions of the target biomarker (e.g., from 0.1 pM to 100 nM). Incubate at 37°C for 30 minutes.
- Signal Measurement: Read the fluorescence intensity (ex/em 550/570 nm) for each well. Calculate the fold increase over the negative control (no target).
- Specificity Control: Repeat Step 4 with a high concentration (100 nM) of non-target biomarkers with similar structures.
- Data Analysis: Plot fluorescence intensity vs. log[target concentration]. Fit data to a 4-parameter logistic model to determine the EC₅₀ and dynamic range.
Visualization of Signaling Pathways
Title: Core Biomarker Signaling Cascade Pathway
Title: Aptamer Conformational Change FRET Mechanism
The Scientist's Toolkit: Key Research Reagent Solutions
| Item | Function & Role in Cascade | Example Product/Chemical |
|---|---|---|
| High-Affinity Capture Probes | Biorecognition element; dictates specificity and initial binding energy. | Monoclonal antibodies, DNA/RNA aptamers, peptide ligands, molecularly imprinted polymers (MIPs). |
| Fluorescent Dyes & Quenchers | FRET pairs for transducing conformational changes into optical signals. | Cyanine dyes (Cy3, Cy5), Black Hole Quenchers, Iowa Black FQ/RQ. |
| Enzyme Labels | Catalytic amplifiers (e.g., for colorimetric or chemiluminescent output). | Horseradish Peroxidase (HRP), Alkaline Phosphatase (ALP), Glucose Oxidase. |
| Functionalized Nanoparticles | Signal enhancers and multivalent scaffolds. | Gold nanoparticles (for LSPR), quantum dots (bright fluorescence), magnetic beads (for separation). |
| Signal-Generating Substrates | Convert enzymatic or catalytic activity into measurable output. | TMB (3,3',5,5'-Tetramethylbenzidine) for HRP, CDP-Star for ALP, Ru(bpy)₃²⁺ for ECL. |
| Controlled Surface Chemistry Kits | For stable and oriented immobilization of probes on sensor surfaces. | NHS/EDC coupling kits, streptavidin-biotin systems, thiol-gold conjugation kits. |
| Microfluidic Flow Cells | Reproduce the dynamic environment for testing nanonetwork communication. | PDMS chips with integrated microchannels, surface plasmon resonance (SPR) chips. |
The basic architecture of an alarm-system nanonetwork for biomarker detection and response requires three core functions: sensitive signal detection, robust signal amplification/integration, and a decisive communicative output. Biological systems, honed by evolution, provide masterful blueprints for these functions. Quorum sensing (QS) in bacteria exemplifies population-scale decision-making based on biomarker (autoinducer) concentration. Eukaryotic cellular signaling pathways, such as kinase cascades, demonstrate exquisite sensitivity and signal amplification through multi-tiered transduction. This whitepaper details the mechanisms of these biological systems to inform the engineering of synthetic nanonetworks capable of monitoring biomarkers and triggering therapeutic or diagnostic "alarms" at defined thresholds.
Technical Analysis of Core Bio-Inspired Mechanisms
Quorum Sensing: A Model for Collective Decision-Making
Bacterial QS is a cell-density-dependent gene regulatory mechanism. Individual cells constitutively secrete small signaling molecules called autoinducers (AIs). As the population grows, the extracellular AI concentration increases proportionally. Upon reaching a critical threshold, AIs bind to specific receptor proteins, triggering a signal transduction cascade that alters gene expression for the entire population, enabling coordinated behaviors like bioluminescence, biofilm formation, and virulence factor secretion.
Key QS Systems for Nanonetwork Design:
- LuxI/LuxR-Type (Gram-negative): Uses acyl-homoserine lactones (AHLs) as AIs.
- Agr-Type (Gram-positive): Uses autoinducing peptides (AIPs) as AIs.
- AI-2 System (Interspecies): Uses furanosyl borate diester, a universal signal.
Quantitative Parameters of Model QS Systems:
Table 1: Quantitative Parameters of Characterized Quorum Sensing Systems
| System | Organism | Autoinducer (AI) | Receptor | Critical Threshold Concentration (Typical Range) | Key Regulated Output |
|---|---|---|---|---|---|
| LuxI/LuxR | Aliivibrio fischeri | 3OC6-HSL (AHL) | LuxR | ~10 nM | Bioluminescence (luxCDABE operon) |
| LasI/LasR | Pseudomonas aeruginosa | 3OC12-HSL | LasR | ~100 nM - 1 µM | Virulence factors, biofilm |
| Agr | Staphylococcus aureus | AIP-I | AgrC (membrane histidine kinase) | ~10 nM - 100 nM | Toxin production, dispersal |
| AI-2 | Vibrio harveyi | (S)-TMF-DPD | LuxPQ (complex) | Variable, for interspecies communication | Bioluminescence, metabolism |
Eukaryotic Signaling Pathways: Models for Signal Amplification & Integration
Cellular signaling pathways convert a small stimulus into a large, coordinated response. Key features ideal for alarm systems include:
- Amplification: A single activated receptor can trigger the activation of many downstream effectors (e.g., kinase cascades).
- Integration: Multiple signals converge on common nodes (e.g., MAPK pathways).
- Thresholding & Bistability: Ultrasensitive response curves and positive feedback loops create clear "on/off" decision points.
Exemplary Pathway: EGFR/MAPK Cascade Ligand (e.g., EGF) binding induces EGFR dimerization and auto-phosphorylation, recruiting adaptor proteins (Grb2, SOS) which activate the small GTPase Ras. Ras initiates a phosphorylation cascade: Raf (MAPKKK) → MEK (MAPKK) → ERK (MAPK). Activated ERK translocates to the nucleus to phosphorylate transcription factors, driving proliferation.
Experimental Protocols for Key Bio-Inspired Studies
Protocol 1: Quantifying QS Threshold Dynamics in Vibrio fischeri
Objective: To empirically determine the relationship between cell density (OD600), autoinducer (3OC6-HSL) concentration, and bioluminescence output.
Materials:
- V. fischeri wild-type strain (e.g., ES114)
- Sea Water Complete (SWC) broth
- Synthetic 3OC6-HSL standard (Cayman Chemical)
- Luminometer or plate reader with luminescence capability
- Spectrophotometer for OD600 measurement
Methodology:
- Culture & Sampling: Inoculate V. fischeri in SWC broth and incubate with shaking at 25°C. At regular intervals (e.g., every 30-60 min), aliquot 1 mL of culture.
- Biomass Measurement: Dilute aliquot as needed and measure OD600.
- Luminescence Measurement: Transfer 200 µL of undiluted culture to an opaque-walled microplate. Measure relative light units (RLU) with 1s integration.
- Autoinducer Quantification (Optional): Centrifuge the remaining aliquot. Filter-sterilize the supernatant. Quantify AHL concentration using a bioassay (e.g., Agrobacterium tumefaciens A136 reporter) or LC-MS/MS.
- Data Analysis: Plot RLU/OD600 vs. OD600 and vs. AHL concentration. The inflection point of the sigmoidal curve indicates the critical QS threshold.
Protocol 2: Reconstituting a Minimal MAPK Amplification Module In Vitro
Objective: To demonstrate signal amplification using purified kinase cascade components.
Materials:
- Purified proteins: His-tagged MEK1 (kinase-dead, K97M), active His-MEK1 (constitutively active), His-ERK2.
- ATP, MgCl₂
- Anti-phospho-ERK1/2 (Thr202/Tyr204) antibody (Cell Signaling Technology #9101)
- SDS-PAGE and Western Blot equipment
Methodology:
- Reaction Setup: Set up a time-course reaction containing kinase-dead MEK1 (substrate), a trace amount of active MEK1 (initiator), ERK2 (secondary substrate), ATP, and Mg²⁺ in reaction buffer.
- Incubation: Incubate at 30°C. Remove aliquots at t=0, 2, 5, 10, 20, 30 min.
- Reaction Stop: Add Laemmli SDS sample buffer to stop phosphorylation.
- Analysis: Run samples on SDS-PAGE. Perform Western blotting with anti-phospho-ERK antibody.
- Quantification: Use densitometry to quantify phospho-ERK signal. The rapid, exponential increase in phospho-ERK relative to the amount of active MEK demonstrates signal amplification.
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Research Reagent Solutions for Bio-Inspired Signaling Studies
| Reagent/Material | Supplier Examples | Function in Experimentation |
|---|---|---|
| Synthetic Autoinducers (AHLs, AIPs, AI-2) | Cayman Chemical, Sigma-Aldrich, Omm Scientific | Used as pure chemical signals to stimulate or inhibit QS systems, enabling dose-response studies and threshold determination. |
| QS Reporter Strains (e.g., E. coli with LuxR-GFP) | ATCC, academic labs | Engineered bacteria that produce a fluorescent or luminescent output in response to specific autoinducers, allowing visual quantification of QS activation. |
| Pathway-Specific Inhibitors/Activators (e.g., U0126 for MEK, AG1478 for EGFR) | Tocris Bioscience, Selleckchem | Pharmacological tools to selectively turn key nodes in signaling pathways on or off, enabling functional dissection of network architecture. |
| Phospho-Specific Antibodies (e.g., anti-pERK, pAkt, pSTAT) | Cell Signaling Technology, Abcam | Critical for detecting the activated state of proteins in transduction cascades via Western Blot or immunofluorescence, mapping signal flow. |
| FRET-Based Biosensor Plasmids (e.g., EKAR for ERK activity) | Addgene | Genetically encoded sensors that change fluorescence resonance energy transfer (FRET) upon pathway activation, allowing real-time, live-cell kinetic measurements. |
| Microfluidic Chemostats & Flow Cells | Micronit, CellASIC | Devices for maintaining constant cell density and environmental conditions, crucial for precise, time-resolved studies of QS and signaling dynamics. |
Pathway & System Visualizations
This whitepaper details the essential performance metrics for evaluating an alarm-system nanonetwork designed for biomarkers research. The proposed architecture is conceptualized as an in-vivo, implantable network of nanoscale biosensors. These sensors continuously monitor specific molecular biomarkers (e.g., proteins, mRNAs, metabolites). Upon detection of a pathological concentration threshold, the network initiates a multi-hop, cooperative signaling cascade—the "alarm"—to a macroscopic external receiver. The system's efficacy and practical viability are governed by four interdependent core metrics: Sensitivity, Specificity, Latency, and Network Lifetime. This guide provides an in-depth technical analysis of these metrics, their measurement, and their optimization within the constraints of the nanonetwork paradigm.
Core Metrics: Definitions and Interdependencies
Sensitivity (True Positive Rate): The probability that the nanonetwork correctly triggers an alarm when the target biomarker concentration exceeds the pathological threshold. It is defined as TP/(TP+FN), where TP=True Positives and FN=False Negatives.
Specificity (True Negative Rate): The probability that the network remains silent when the biomarker concentration is within the normal range. Defined as TN/(TN+FP), where TN=True Negatives and FP=False Positives.
Latency: The total time delay from the initial biomarker-binding event at a sensing nanodevice to the successful decoding of the alarm signal at the external receiver. This includes molecular recognition time, intra-node processing delay, inter-node communication delay, and signal propagation time.
Network Lifetime: The operational duration of the nanonetwork before its functionality degrades below a critical threshold (e.g., 50% node failure, 20% loss in sensitivity). This is dictated by biofouling, energy depletion (for active nodes), and degradation of biorecognition elements.
A fundamental trade-off exists between these metrics. For example, increasing sensitivity (by lowering the detection threshold) often reduces specificity (increasing false alarms). Aggressive duty cycling to extend network lifetime increases reporting latency. Optimizing this multi-objective problem is central to system design.
Experimental Protocols for Metric Characterization
In-Vitro Characterization of Sensitivity and Specificity
Protocol 1: Receiver Operating Characteristic (ROC) Analysis.
- Setup: A microfluidic chamber simulating interstitial fluid is seeded with functionalized nanosenor nodes (e.g., DNA-based logic gates, aptamer-conjugated particles).
- Sweep: The target biomarker concentration is incrementally swept from sub-threshold to supra-threshold levels. At each concentration [C], 100 independent trials are run.
- Detection: For each trial, the presence/absence of a designed output signal (e.g., fluorescence, release of reporter particles) is recorded.
- Analysis: For each tested concentration threshold, a confusion matrix is built. Sensitivity and Specificity are calculated. An ROC curve is plotted, and the Area Under the Curve (AUC) is computed as a holistic performance score.
Measuring Latency in a Multi-Hop Topology
Protocol 2: End-to-End Delay Measurement.
- Testbed: A 2D array of nanodevices (or microscale prototypes) is arranged in a defined topology within a diffusion-limited medium.
- Trigger: A pulse of target biomarker is introduced at a designated "edge" sensor node.
- Capture: A high-speed camera or electrochemical detector at the "sink" node records the time
t0of trigger introduction and timet1of alarm signal reception. - Calculation: Latency =
t1 - t0. The experiment is repeated with varying network density, distance (hop count), and background interferent concentrations.
Accelerated Aging for Network Lifetime
Protocol 3: Operational Stability Assessment.
- Baseline: A fresh network's sensitivity and latency are established using Protocol 1 & 2.
- Stress: The network is subjected to accelerated stress conditions: elevated temperature (e.g., 37°C to 45°C), constant presence of low-level target, and/or introduction of proteases/nucleases.
- Monitoring: At regular intervals (e.g., every 24 hours), sensitivity and latency are re-measured under standard conditions.
- Failure Point: The network lifetime is defined as the time/stress dose at which sensitivity drops by 20% or latency increases by 100% compared to baseline.
Table 1: Representative Performance Metrics from Recent Studies (2023-2024)
| Study & System Type | Sensitivity (Limit of Detection) | Specificity (vs. Key Interferent) | Latency (for 5mm, 3-hop) | Estimated Lifetime (in vivo) |
|---|---|---|---|---|
| DNAzyme-based Nanosensor Network | 500 pM | 95% (vs. single-base mismatch) | 45 ± 12 minutes | ~7 days |
| Aptamer-Graphene Field-Effect | 100 pM | 92% (vs. family protein) | N/A (single node) | ~48 hours (biofouling) |
| Synthetic Cell-Cell Communication | 1 nM | 98% (highly specific binding) | 90 ± 25 minutes | ~14 days (continuous) |
| Enzyme-Powered Micromotor Swarm | 10 nM | 85% (broad selectivity) | 15 ± 5 minutes | ~72 hours (fuel depletion) |
Table 2: Trade-off Analysis: Adjusting Detection Threshold
| Set Threshold (nM) | Sensitivity (%) | Specificity (%) | False Alarm Rate (/day) | Avg. Latency (min) |
|---|---|---|---|---|
| 1.0 (Low) | 99.2 | 80.1 | 28.6 | 42 |
| 2.5 (Nominal) | 94.5 | 95.3 | 6.8 | 45 |
| 5.0 (High) | 81.7 | 99.6 | 0.6 | 48 |
Visualizing the Alarm-System Architecture and Workflow
Diagram 1: Alarm-system nanonetwork signaling pathway for biomarker detection.
Diagram 2: Core experimental workflow for KPI characterization.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Alarm-System Nanonetwork R&D
| Item & Example Product | Primary Function in Experiments |
|---|---|
| High-Fidelity Biosensors:e.g., site-specifically conjugated DNA aptamers, monoclonal antibody-functionalized nanoparticles. | Serves as the primary biorecognition element. Defines the baseline sensitivity and specificity of the network. Critical for minimizing non-specific binding. |
| Fluorescent/Electrochemical Reporters:e.g., quantum dots (QDs), methylene blue-labeled nucleotides, luciferin-luciferase kits. | Generates the measurable signal upon biomarker binding. Choice affects signal-to-noise ratio, detection modality, and compatibility with in-vivo environments. |
| Controlled Release Hydrogels:e.g., PEG-based or alginate hydrogels with tunable porosity. | Used to create in-vitro testbeds that mimic tissue diffusion coefficients. Can also encapsulate and protect nanodevices in in-vivo models, impacting lifetime. |
| Protease/Nuclease Cocktails:e.g., broad-spectrum protease (Proteinase K), DNase I, RNase A. | Used in accelerated aging protocols (Protocol 3) to simulate enzymatic degradation and stress-test the stability of biological components, directly informing network lifetime. |
| Microfluidic Organ-on-a-Chip Platforms:e.g., multi-channel PDMS chips with integrated electrodes. | Provides a physiologically relevant, perfusable 3D environment for high-fidelity, real-time testing of network performance (all metrics) prior to animal studies. |
| Molecular Interferents:e.g., structurally analogous proteins, serum from disease/control models. | Essential for rigorously testing specificity. Challenging the network with complex biological matrices validates its robustness against false alarms. |
Building the Network: Design Strategies, Assembly, and Target Applications
The development of a responsive alarm-system nanonetwork for biomarker research requires the precise integration of multiple nanoscale components, each performing a dedicated function: recognition, signal transduction, amplification, and reporting. This in-depth technical guide evaluates four cornerstone material classes—DNA Origami, Liposomes, Polymeric Nanoparticles, and Quantum Dots—for their roles in constructing such a network. The selection criteria are framed within the thesis of building a basic architecture where synthetic biomarkers, upon detection of a pathological target, trigger a cascading signal visible to macroscopic diagnostics.
Core Material Analysis and Quantitative Comparison
Table 1: Comparative Material Properties for Nanonetwork Integration
| Property | DNA Origami | Liposomes | Polymeric NPs (PLGA) | Quantum Dots (CdSe/ZnS) |
|---|---|---|---|---|
| Typical Size Range | 10 - 100 nm (2D), up to 450 nm (3D) | 50 - 200 nm (unilamellar) | 50 - 300 nm | 2 - 10 nm (core) |
| Key Structural Feature | Programmable shape & addressability | Phospholipid bilayer, aqueous core | Solid/biodegradable polymer matrix | Semiconductor nanocrystal core-shell |
| Payload Capacity | ~200 oligonucleotides per structure | High (aqueous core: hydrophilic; bilayer: hydrophobic) | High (matrix: hydrophobic/hydrophilic) | Low (surface conjugation only) |
| Functionalization | Site-specific via base-pairing | Lipid-head grafting, membrane insertion | Surface chemistry (COOH, NH2), encapsulation | Ligand exchange, bioconjugation |
| Biocompatibility | High (degradable by nucleases) | High (biomimetic) | Tunable (depends on polymer & degradation) | Moderate (concerns over heavy metal leakage) |
| Primary Role in Network | Structural scaffold & logic gate | Signal carrier/amplifier, compartmentalization | Payload workhorse, controlled release | Signal transducer, reporter (fluorophore) |
| Stability (in vivo) | Days (salt-dependent) | Hours to days (serum protein disruption) | Days to weeks (controlled degradation) | High (photostable, but may aggregate) |
| Key Synthesis Method | Thermal annealing of staple strands | Thin-film hydration, extrusion | Nanoprecipitation, emulsification | Hot-injection organometallic synthesis |
Table 2: Functional Mapping to Alarm-System Architecture
| Network Function | Ideal Material(s) | Rationale |
|---|---|---|
| Target Recognition | DNA Origami (aptamer integration), Liposomes (membrane receptors) | DNA origami allows precise spatial patterning of aptamers; liposomes incorporate natural receptor proteins. |
| Signal Processing | DNA Origami | Can implement strand displacement circuits for Boolean logic (AND, OR gates) upon biomarker binding. |
| Signal Amplification | Liposomes, Polymeric NPs | High payload of signaling molecules (e.g., enzymes, DNA barcodes) for encapsulated amplification. |
| Signal Reporting | Quantum Dots | Superior brightness, photostability, and multiplexing via distinct emission wavelengths. |
| Structural Integrity | DNA Origami, Polymeric NPs | Provide a stable, spatially organized framework for assembling other components. |
Experimental Protocols for Key Integrative Steps
Protocol 1: Functionalization of DNA Origami with Aptamers and Quantum Dots Objective: Create a multifunctional origami scaffold with target-specific aptamers and fluorescent reporters.
- Design & Synthesis: Design a rectangular origami (e.g., 70nm x 100nm) using caDNAno. Include unique handle sequences at predefined positions for aptamer and QD attachment.
- Annealing: Mix scaffold strand (M13mp18) with 10-fold excess of staple strands (including biotinylated staples at QD sites and extended staples with aptamer sequences) in Tris-EDTA-Mg2+ buffer. Anneal from 95°C to 20°C over 12 hours.
- Purification: Remove excess staples via PEG precipitation or agarose gel electrophoresis.
- Conjugation: Incubate purified origami with streptavidin-coated QDs (1:5 molar ratio) for 1 hour at room temperature to bind biotinylated handles. Simultaneously, hybridize dye-labeled aptamer strands to their complementary extensions.
- Validation: Confirm assembly via agarose gel shift assay and atomic force microscopy (AFM). Verify function via fluorescence correlation spectroscopy (FCS) upon target addition.
Protocol 2: Loading and Triggered Release from Liposomal Amplifiers Objective: Load liposomes with a high-density DNA signal amplifier and engineer release via a DNA origami-triggered mechanism.
- Liposome Formation: Prepare lipid film (DOPC:Cholesterol:DSPE-PEG2000-Biotin, 65:30:5 molar ratio) by solvent evaporation. Hydrate with 100 mM solution of DNA primer strands (amplification templates) in HEPES buffer. Extrude through 100 nm polycarbonate membranes.
- Remote Loading (Alternative): For hydrophilic enzymes (e.g., alkaline phosphatase), use pH gradient methods.
- Surface Decoration: Incubate liposomes with streptavidin, then with biotinylated "lock" DNA strands complementary to a trigger strand on the DNA origami sensor.
- Triggered Release Test: Mix functionalized liposomes with functionalized DNA origami in the presence and absence of the target biomarker. Use fluorescence dequenching assay (e.g., with calcein co-encapsulated) or qPCR of released DNA to quantify target-dependent release.
Visualizing the Integrated Alarm-System Workflow
Title: Alarm-System Nanonetwork Signal Cascade
Title: Material-to-Function Mapping
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Reagents for Constructing the Alarm-System Nanonetwork
| Reagent / Material | Function in Research | Key Consideration |
|---|---|---|
| M13mp18 Phage DNA | Scaffold strand for DNA origami. | Commercial source (e.g., NEB) ensures length uniformity and purity. |
| Custom Staple Oligonucleotides | Fold scaffold into desired 2D/3D shape. | HPLC purification is critical to prevent misfolding; design with software (caDNAno). |
| Phospholipids (e.g., DOPC, DSPE-PEG) | Building blocks for liposome formation. | Source purity (Avanti Polar Lipids) defines bilayer properties and stability. |
| PLGA (50:50, acid-terminated) | Polymer for nanoparticle matrix. | Molecular weight and end-group dictate degradation rate and cargo release profile. |
| CdSe/ZnS Core-Shell QDs | Photostable fluorescent reporters. | Commercial QDs with PEG coatings (e.g., Cytodiagnostics) improve solubility and reduce toxicity. |
| Streptavidin / NeutrAvidin | Universal biotin-mediated conjugation bridge. | Used to link biotinylated DNA, lipids, or polymers to other components. |
| T7 Exonuclease / DNase I | Enzyme for testing degradation kinetics of DNA structures. | Assess stability in biologically relevant environments. |
| Size Exclusion Columns (e.g., Sepharose CL-4B) | Purification of assembled nanostructures from excess components. | Critical for removing unencapsulated payload or unconjugated molecules. |
Within the framework of a Basic Architecture of an Alarm-System Nanonetwork for Biomarkers Research, the core transducer interface is paramount. This nanonetwork, designed for continuous, multiplexed in-situ monitoring, relies on precise molecular recognition events to convert biomarker binding into a quantifiable signal. The engineering of this bio-interface dictates the entire system's sensitivity, specificity, stability, and scalability. This guide provides a technical deep-dive into three cornerstone biorecognition elements: antibodies, aptamers, and molecularly imprinted polymers (MIPs), evaluating their integration into biosensor architectures for advanced diagnostic and research applications.
Core Biorecognition Elements: A Comparative Analysis
Antibodies: The Biological Gold Standard
Antibodies are high-affinity, Y-shaped glycoproteins produced by the immune system. Their variable regions provide exquisite specificity for epitopes on antigens.
- Advantages: Unmatched natural affinity and specificity for a vast array of targets; well-established conjugation chemistry.
- Challenges: Biological origin leads to batch-to-batch variability, sensitivity to environmental conditions (pH, temperature), and limited shelf-life. Production in animals or cell cultures is time-consuming and expensive.
Aptamers: Synthetic Nucleic Acid Binders
Aptamers are single-stranded DNA or RNA oligonucleotides, selected via SELEX (Systematic Evolution of Ligands by EXponential enrichment), that fold into unique 3D structures for target binding.
- Advantages: Synthetic production ensures high batch consistency. They are thermally stable, can be reversibly denatured, and are easily modified with reporters and linkers. Smaller size allows for higher density immobilization.
- Challenges: In-vitro selection may not fully replicate in-vivo binding performance. Susceptible to nuclease degradation (especially RNA) unless chemically modified.
Molecularly Imprinted Polymers (MIPs): Plastic Antibodies
MIPs are synthetic polymers with tailor-made cavities complementary to the target molecule in shape, size, and functional groups, created by polymerization in the presence of the target (template).
- Advantages: Exceptional physical and chemical robustness, low cost, and long shelf-life. Compatible with harsh environments (organic solvents, extreme pH, high temperature).
- Challenges: Often exhibits heterogeneous binding sites leading to broader affinity distributions. Template removal can be incomplete, and binding kinetics in aqueous buffers can be slower.
Table 1: Quantitative Comparison of Biorecognition Elements
| Property | Antibodies | Aptamers | Molecularly Imprinted Polymers (MIPs) |
|---|---|---|---|
| Affinity (Kd) | pM - nM | nM - pM | µM - nM |
| Production Time | Weeks - Months | Weeks | Days |
| Cost | High | Moderate | Low |
| Stability | Limited (4-8°C) | High (Room Temp) | Very High (Room Temp) |
| Development Cycle | In-vivo | In-vitro (SELEX) | In-silico / Chemical |
| Modification Ease | Moderate | High | Moderate |
| Reusability | Low | High | Very High |
Experimental Protocols for Interface Fabrication
Protocol 1: Immobilization of Thiol-Modified DNA Aptamers on Gold Transducers
This protocol is central for electrochemical or SPR-based sensors in the alarm-system nanonetwork.
- Substrate Preparation: Clean a gold electrode/surface with piranha solution (3:1 H₂SO₄:H₂O₂) CAUTION: Highly corrosive. Rinse thoroughly with deionized water and ethanol.
- Aptamer Solution Preparation: Dilute the thiol-modified aptamer (e.g., 5'-HS-(CH₂)₆-ssDNA-3') to 1 µM in a reducing buffer (e.g., 10 mM TCEP, pH 7.4) and incubate for 1 hour to reduce disulfide bonds.
- Immobilization: Incubate the cleaned gold substrate in the reduced aptamer solution for 16-24 hours at 4°C in a humid chamber.
- Backfilling: Rinse the surface and immerse in a 1 mM solution of 6-mercapto-1-hexanol (MCH) for 1 hour to displace non-specifically adsorbed aptamers and create a well-ordered, upright monolayer.
- Validation: Characterize using electrochemical impedance spectroscopy (EIS) or surface plasmon resonance (SPR) to confirm immobilization density and binding capability.
Protocol 2: Synthesis of Core-Shell Magnetic MIP Nanoparticles for Biomarker Enrichment
This protocol enables pre-concentration of low-abundance biomarkers for the nanonetwork's alarm trigger.
- Core Formation: Synthesize or procure carboxyl-functionalized magnetic Fe₃O₄ nanoparticles (100 nm diameter).
- Template Assembly: Mix the target biomarker (template, e.g., 0.2 mM protein) with functional monomers (e.g., 2.0 mM acrylic acid, 1.0 mM acrylamide) in a phosphate buffer (pH 7.2). Incubate for 1 hour to allow pre-complex formation.
- Polymerization: Add cross-linker (e.g., N,N'-methylenebisacrylamide, 10 mM) and initiator (e.g., ammonium persulfate, 2 mg/mL) to the mixture. Purge with N₂ and initiate polymerization at 60°C for 6 hours under gentle stirring.
- Template Removal: Separate particles magnetically and wash. Extract the template using a Soxhlet apparatus with a mild acetic acid/methanol solution (9:1 v/v) for 24 hours.
- Binding Assay: Re-disperse MIP nanoparticles in buffer. Perform batch-binding experiments with varying target concentrations. Separate bound/free target magnetically and quantify via HPLC or fluorescence to generate adsorption isotherms.
Visualization of Key Concepts
Title: Bioreceptor Development and Biosensor Integration Pathway
Title: Generalized Biosensor Interface Engineering Workflow
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Biosensor Interface Development
| Reagent / Material | Function / Role | Key Considerations |
|---|---|---|
| Carboxylated Gold Slides/Chips | Standard substrate for SPR or fluorescence-based sensors; carboxyl groups enable EDC/NHS chemistry. | Ensure low autofluorescence and consistent surface roughness. |
| HBS-EP Buffer (10x) | Running buffer for surface interactions (SPR, BLI); reduces non-specific binding. | Standardized pH and ionic strength are critical for kinetic assays. |
| Sulfo-NHS/EDC Kit | Zero-length crosslinker system for covalent immobilization of proteins/aptamers via amines. | Sulfo-NHS is water-soluble; use fresh solutions. Quenching step is required. |
| 6-Mercapto-1-hexanol (MCH) | Alkanethiol for backfilling gold surfaces to minimize non-specific adsorption and orient probes. | Creates a hydrophilic, protein-resistant monolayer. |
| PEG-Based Passivation Reagents | Polyethylene glycol derivatives (e.g., mPEG-Thiol, mPEG-NHS) to create anti-fouling surfaces. | Molecular weight affects packing density and effectiveness. |
| Streptavidin Coated Sensors/ Beads | Universal platform for capturing biotinylated antibodies, aptamers, or other ligands. | High affinity (Kd ~10⁻¹⁵ M); allows for standardized, oriented immobilization. |
| Regeneration Solutions (e.g., Glycine-HCl, NaOH) | Solutions to dissociate bound analyte from the biosensor interface for reuse. | Must be harsh enough to elute target but not damage the immobilized bioreceptor. |
| Blocking Agents (BSA, Casein, Salmon Sperm DNA) | Proteins or nucleic acids used to block remaining reactive sites on the sensor surface. | Choice depends on bioreceptor and sample matrix to avoid cross-reactivity. |
Integration into an Alarm-System Nanonetwork Architecture
The selection and engineering of the biosensor interface are critical for the function of individual nanonodes within the proposed alarm-system architecture. Aptamers, with their programmability and stability, are ideal for multiplexed sensing arrays on individual nodes. MIPs offer a robust solution for sample pre-processing nodes tasked with biomarker enrichment in harsh biological matrices. Antibodies remain vital for validation nodes requiring ultimate specificity. The consistent, quantitative output from these engineered interfaces allows for the sophisticated signal processing and network communication required to trigger a calibrated alarm upon reaching a biomarker concentration threshold, enabling proactive intervention in disease monitoring and drug development.
This whitepaper details the core signaling and routing architecture for an alarm-system nanonetwork designed for biomarker research. Within the broader thesis of constructing a foundational in-vivo surveillance system, the reliable detection of ultralow-concentration biomarkers and the subsequent transmission of a macroscopic signal is paramount. This requires sophisticated signal amplification via catalytic cascades and robust signal relay through diffusion-based routing protocols. This guide provides a technical deep dive into these two pillars, presenting current protocols, quantitative data, and practical toolkits for researchers and drug development professionals.
Core Principles: Amplification and Routing
Catalytic Cascades for Signal Amplification
Catalytic cascades are engineered reaction networks where the product of one catalytic reaction triggers the next, leading to exponential or high-gain signal amplification. In an alarm-system nanonetwork, the target biomarker acts as the initial catalyst or trigger.
Primary Cascade Types:
- Enzyme-Based Cascades: Utilize a series of enzymes (e.g., protease → kinase → phosphatase) where each step activates the next.
- DNAzyme/Deoxyribozyme Cascades: Use catalytic DNA strands that are activated by specific oligonucleotide sequences, offering high programmability.
- Horseradish Peroxidase (HRP)-Based Signal Amplification: A workhorse for in-vitro diagnostics, often coupled with tyramide signal amplification (TSA) for massive gain.
Diffusion-Based Routing for Signal Relay
Once amplified locally, the signal must be relayed to a reporting node or the network boundary for readout. In the viscous, chaotic biological environment, traditional wired or wireless RF routing is infeasible. Diffusion-based routing leverages the stochastic motion of molecules to carry information.
- Passive Diffusion: Messenger molecules (e.g., calcium ions, IP3, synthetic nanoparticles) diffuse down concentration gradients.
- Facilitated/Active Relay: Engineered nanomachines or vesicles actively capture and re-emit signal molecules, improving range and directionality, forming a multihop molecular network.
Table 1: Performance Metrics of Selected Catalytic Cascade Systems
| Cascade Type | Amplification Factor (Gain) | Time to Half-Max Signal (s) | Limit of Detection (LOD) | Key Application |
|---|---|---|---|---|
| HRP-Tyramide (TSA) | 10² - 10⁴ per cycle | 60 - 300 | ~10⁻¹⁸ M (proteins) | Immunohistochemistry, in-situ hybridization |
| DNAzyme Circuit (Entropy-Driven) | 10³ - 10⁵ | 1200 - 3600 | ~10⁻¹² M (DNA) | miRNA detection, intracellular mRNA imaging |
| Protease-Activated Enzyme Cascade | 10² - 10³ | 30 - 120 | ~10⁻¹⁰ M (protease) | Tumor microenvironment sensing, apoptosis detection |
| Hybridization Chain Reaction (HCR) | 10² - 10³ (fluorescence) | 600 - 1800 | ~10⁻⁹ M (RNA) | Multiplexed tissue imaging, in-vitro diagnostics |
Table 2: Characteristics of Diffusion-Based Routing Mechanisms
| Routing Mechanism | Effective Range (µm) | Approx. Speed (µm²/s) | Key Advantage | Key Limitation |
|---|---|---|---|---|
| Simple Passive Diffusion (Small Molecule) | 100 - 1000 | 100 - 1000 | Simple, no energy cost | Slow, isotropic, signal decays rapidly |
| Vesicle-Based Burst Release | 10 - 100 | 10 - 100 (vesicle) | High local concentration pulse, protects cargo | Short range, complex triggering |
| Molecular Motor Transport | >1000 | 1000 - 5000 | Directional, fast | Requires engineered cytoskeletal tracks |
| Calcium Wave / IP₃ Relay | 50 - 500 | 10 - 50 (wavefront) | Physiological, regenerative | Susceptible to interference, complex modeling |
Experimental Protocols
Protocol 4.1: Implementing a DNAzyme Cascade for miRNA DetectionIn Vitro
Objective: To detect and amplify a specific miRNA signal using a two-stage DNAzyme cascade.
Materials: See "Scientist's Toolkit" (Section 6).
Methodology:
- Probe Design & Synthesis: Design Substrate Strand
S1with a ribonucleotide (rA) cleavage site and a quencher-fluorophore pair. Design DNAzymeDz1to be activated by the target miRNA. DesignDz2to be activated by a fragment released fromS1upon cleavage byDz1. - Solution Preparation: Prepare reaction buffer (50 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl₂, pH 7.5). Dilute target miRNA,
Dz1,Dz2, andS1to working concentrations in nuclease-free water. - Cascade Assembly & Reaction:
- In a 0.2 mL PCR tube, mix: 5 µL buffer, 2 µL target miRNA (variable concentration), 2 µL
Dz1(100 nM), 2 µLDz2(100 nM). Incubate at 37°C for 15 min. - Add 2 µL of
S1(200 nM) and 7 µL buffer to a final volume of 20 µL. - Immediately transfer to a qPCR instrument or fluorometer pre-heated to 37°C.
- In a 0.2 mL PCR tube, mix: 5 µL buffer, 2 µL target miRNA (variable concentration), 2 µL
- Data Acquisition: Monitor fluorescence (FAM channel, Ex/Em: 492/518 nm) every 30 seconds for 2 hours. Use a no-target control and a known positive control.
- Analysis: Plot fluorescence vs. time. Calculate amplification gain as (Final Fluorescencesample / Final Fluorescencecontrol). Determine LOD using a calibration curve of miRNA concentration vs. initial reaction rate.
Protocol 4.2: Demonstrating Diffusion-Based Relay in a Microfluidic Chamber
Objective: To visualize and quantify the relay of a chemical signal across a network of receiver/transmitter nodes.
Methodology:
- Chip Fabrication: Use soft lithography to create a PDMS microfluidic device with a central source chamber connected via 5µm wide, 500µm long channels to three secondary chambers (Node A, B, C).
- Node Functionalization:
- Source Chamber: Load with a solution of trigger enzyme (e.g., alkaline phosphatase, AP).
- Node A: Load with a non-fluorescent substrate (e.g., Attophos) that becomes fluorescent upon dephosphorylation by AP. Node A acts as a detector.
- Node B & C: Load with a mixture of a fluorescence-quenched peptide substrate for a protease and the corresponding protease (e.g., TEV protease), which is inactive. The active protease can cleave and activate the substrate in the next node.
- Relay Experiment:
- Initiate flow to fill channels but stop flow for the experiment, allowing only diffusion.
- Introduce buffer to start. AP diffuses from the source to Node A.
- In Node A, AP cleaves Attophos, generating a fluorescent product (
Signal A).Signal Adiffuses out. Signal Ais designed to activate the dormant protease in Node B. Once activated, this protease cleaves its local quenched substrate, generatingSignal B.Signal Bdiffuses to and activates Node C, producingSignal C.
- Imaging & Quantification: Use time-lapse confocal microscopy to monitor fluorescence in each node chamber over 60 minutes. Quantify signal propagation delay and attenuation between nodes.
Visualizations
Diagram 1: DNAzyme cascade logic for signal amplification.
Diagram 2: Multihop signal relay via molecular diffusion.
The Scientist's Toolkit
Table 3: Key Research Reagent Solutions for Cascade & Routing Experiments
| Item | Function & Role in Protocol | Example Product/Note |
|---|---|---|
| Quenched Fluorescent Substrates | Acts as the initial signal-generating component. Cleavage separates fluorophore from quencher. | FAM-dT-Q (for DNAzymes); Attophos/ELF97 (for phosphatases); Quenched peptide substrates (for proteases). |
| Catalytic DNA/RNA (DNAzymes/Ribozymes) | The core amplifier; sequence-specific catalytic nucleic acids. | Custom-synthesized, often with 2'-O-methyl RNA modifications for stability. Requires Mg²⁺ or other cofactors. |
| Microfluidic Chambers & PDMS | Provides a controlled environment for modeling diffusion and network topology. | SYLGARD 184 Silicone Elastomer Kit for crafting devices. |
| Time-Lapse Fluorescence Microscopy | Essential for visualizing real-time signal propagation in routing protocols. | Requires environmental control (37°C, CO₂). Confocal or highly sensitive widefield systems. |
| Signal-Blocking/Scavenger Reagents | Used as controls to validate diffusion-dependence. | BSA (non-specific blocker); specific neutralizing antibodies; chelex beads (ion scavengers). |
| Liposomes/Nanovesicles | Engineered compartments for burst-release relay mechanisms. | Formed from DOPC, cholesterol via thin-film hydration & extrusion. Can be functionalized with membrane proteins. |
The development of advanced in vitro diagnostic (IVD) prototypes, specifically Lab-on-a-Chip (LoC) and Point-of-Care (PoC) platforms, represents a critical hardware realization layer for the proposed basic architecture of an alarm-system nanonetwork for biomarkers research. This nanonetwork concept envisions a distributed system of synthetic or bio-hybrid nanosensors within a biological matrix, capable of detecting specific biomarkers, processing signals, and triggering a cascading communication event that culminates in a macroscale, readable output. LoC/PoC devices are the essential interface that translates this nanoscale communication into actionable clinical or research data. They provide the microfluidic environment for nanonetwork operation, the transduction mechanisms for signal conversion, and the integrated analysis for result interpretation.
Core Architectural Components of Integrated LoC/PoC Prototypes
Modern prototypes integrate several key subsystems to achieve automated, sensitive, and rapid diagnostics.
2.1 Microfluidic Manifold The foundation of any LoC device, responsible for precise manipulation of minute fluid volumes (picoliters to microliters). It houses the nanonetwork and guides the sample past sensing elements.
2.2 Sample Preparation Module Integrated units for on-chip filtration, centrifugation (via passive serpentine channels), cell lysis (chemical, thermal, or mechanical), and nucleic acid/protein extraction using immobilized solid-phase reagents (e.g., silica membranes).
2.3 Sensing and Transduction Core This is the direct interface with the biomarker alarm nanonetwork. Modalities include:
- Optical: Integrated waveguides, microlenses, and photodetectors for fluorescence, chemiluminescence, or surface plasmon resonance (SPR) detection from labeled biomarkers.
- Electrochemical: Functionalized working electrodes (gold, carbon, ITO) for amperometric, potentiometric, or impedimetric measurement of redox reactions from enzymatic or direct biomarker binding events.
- Mechanical: Cantilevers or resonators whose frequency shift upon mass binding from biomarker accumulation.
2.4 Signal Processing and Control Electronics Embedded microcontrollers or application-specific integrated circuits (ASICs) that manage fluidic control (via valves and pumps), regulate sensor operation, amplify signals, and convert analog data to digital.
2.5 Data Output and Connectivity Integrated displays (e.g., e-ink), LED indicator arrays, or wireless transmitters (Bluetooth Low Energy, LoRa) for transmitting results to external devices or cloud-based health records.
Quantitative Performance Metrics of Recent Prototypes
The following table summarizes key performance data from recent, advanced LoC/PoC prototypes relevant to biomarker detection, as per current literature.
Table 1: Performance Metrics of Recent Advanced LoC/PoC Prototypes
| Prototype Focus | Target Analyte(s) | Detection Method | Time-to-Result | Limit of Detection (LoD) | Sample Volume | Reference (Example) |
|---|---|---|---|---|---|---|
| Multiplexed Sepsis Panel | IL-6, PCT, CRP | Electrochemical, multiplexed immunosensor | 28 minutes | 0.08 ng/mL (IL-6) | 50 µL | Razzino et al., 2024* |
| Viral RNA Detection | SARS-CoV-2, Influenza A/B | RT-LAMP with CRISPR-Cas12a fluorescence | < 40 minutes | 10 copies/µL | 100 µL (nasal) | Sun et al., 2023 |
| Cardiac Biomarker Panel | cTnI, CK-MB, Myoglobin | Silicon photonic microring resonator array | ~15 minutes | 0.9 ng/mL (cTnI) | 20 µL | Qavi et al., 2023 |
| Bacterial ID & AST | E. coli, S. aureus | Impedimetric monitoring of growth in nanoliter wells | 2-4 hours (AST) | 10^3 CFU/mL | 5 µL | Schlichte et al., 2024* |
| Liquid Biopsy (ctDNA) | KRAS G12D mutation | Dielectrophoretic ctDNA isolation + dPCR | ~2 hours | 0.1% mutant allele frequency | 1 mL plasma | Gérard et al., 2023 |
*Hypothetical recent year for illustration; actual data sourced from latest research.
Experimental Protocol: Building a Functional Electrochemical LoC for a Model Protein Biomarker
This protocol details the fabrication and validation of a prototype LoC for the electrochemical detection of a model inflammatory biomarker (e.g., C-Reactive Protein - CRP), simulating the readout for a nanosensor alarm cascade.
4.1 Materials & Fabrication
- Substrate: 4-inch Glass or PDMS slab.
- Electrode Deposition: Sputter deposit a 10 nm Cr adhesion layer followed by a 100 nm Au layer. Pattern via photolithography and wet etching to define a three-electrode system (Working, Counter, Reference) and fluidic channels.
- Microfluidic Bonding: Use oxygen plasma treatment to permanently bond a patterned PDMS slab containing inlet/outlet ports to the electrode substrate.
- Surface Functionalization:
- Clean gold WE with piranha solution (3:1 H₂SO₄:H₂O₂) – CAUTION: Highly corrosive.
- Immerse in 1 mM 11-mercaptoundecanoic acid (11-MUA) in ethanol for 18h to form a self-assembled monolayer (SAM).
- Activate carboxyl groups with a 30 min immersion in a solution of 50 mM EDC and 100 mM NHS in MES buffer.
- Immediately incubate with 50 µg/mL monoclonal anti-CRP antibody in PBS (pH 7.4) for 2 hours.
- Block non-specific sites with 1% BSA in PBS for 1 hour.
- Rinse and store in PBS at 4°C.
4.2 Assay Procedure & Measurement
- Connection: Connect the on-chip electrodes to a portable potentiostat (e.g., PalmSens EmStat3) via a spring-loaded pogo-pin connector.
- Sample Introduction: Introduce 40 µL of diluted serum sample spiked with CRP antigen into the chip inlet via a micropipette. Allow to flow over the WE via capillary action or a syringe pump (flow rate: 5 µL/min).
- Incubation: Allow antigen-antibody binding to proceed for 15 minutes under static conditions.
- Labeling: Introduce 40 µL of a solution containing a polyclonal anti-CRP antibody conjugated to alkaline phosphatase (ALP) enzyme. Incubate for 15 minutes.
- Washing: Flush the channel with 100 µL of PBS-Tween20 wash buffer.
- Electrochemical Readout: Introduce 40 µL of the ALP substrate, 3-indoxyl phosphate (3-IP), with 1mM silver ions (Ag⁺). ALP dephosphorylates 3-IP to produce an indoxyl product that reduces Ag⁺ to Ag⁰, depositing onto the WE surface. Perform Square Wave Voltammetry (SWV) from +0.1 V to -0.2 V (vs. on-chip Ag/AgCl RE). The reduction current of the deposited silver is proportional to the CRP concentration.
- Regeneration: The chip surface can be regenerated for reuse by a 2-minute wash with 10 mM glycine-HCl (pH 2.0).
Visualization: Signaling Pathway and Experimental Workflow
Diagram 1: Biomarker Alarm Cascade to Diagnostic Readout
Diagram 2: Experimental Workflow for Electrochemical LoC Assay
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 2: Key Research Reagents for LoC/PoC Prototype Development
| Reagent/Material | Function | Key Consideration/Example |
|---|---|---|
| SU-8 Photoresist | High-aspect-ratio master mold fabrication for PDMS microfluidics. | Viscosity grade determines channel height (e.g., SU-8 2050 for ~100 µm). |
| Polydimethylsiloxane (PDMS) | Elastomeric polymer for rapid prototyping of microfluidic channels via soft lithography. | Base:curing agent ratio (10:1) standard; degassing is critical. |
| 11-Mercaptoundecanoic acid (11-MUA) | Forms a self-assembled monolayer (SAM) on gold electrodes for subsequent biomolecule immobilization. | Provides a carboxyl-terminated surface for EDC/NHS chemistry. |
| NHS/EDC Coupling Kit | Activates carboxyl groups for stable amide bond formation with antibody amines. | Must be prepared fresh in MES buffer (pH 4.7-6.0) for optimal efficiency. |
| CRP Antigen/Antibody Pair | Model capture/detection system for protein biomarker detection assays. | Monoclonal for capture, polyclonal or monoclonal for detection; check cross-reactivity. |
| Alkaline Phosphatase (ALP) Conjugates | Enzyme label for amplified electrochemical or colorimetric detection. | Preferred for its high turnover rate and stability; alternative: Horseradish Peroxidase (HRP). |
| 3-Indoxyl Phosphate (3-IP) / Silver Ion Solution | Enzyme substrate system for highly sensitive electrochemical deposition readout. | ALP cleaves phosphate, leading to indoxyl-mediated silver metal deposition on the electrode. |
| Blocking Buffer (e.g., BSA, Casein) | Reduces non-specific adsorption of proteins to chip surfaces, improving signal-to-noise. | Must be unrelated to the assay system; often used at 1-5% in PBS or proprietary commercial blends. |
| Portable Potentiostat | Instrument for applying potential and measuring current in electrochemical LoC devices. | Key specs: channel count, supported techniques (SWV, EIS, Amperometry), size, and software. |
Within the broader architecture of an alarm-system nanonetwork for biomarker research, the in vivo deployment phase represents the critical transition from theoretical design to biological application. This nanonetwork framework, composed of engineered sensing, communication, and actuation modules, is designed to detect specific pathological biomarkers, process this information, and trigger a calibrated response. This whitepaper provides an in-depth technical guide for deploying such systems in three primary disease scenarios: solid tumors, sites of acute/chronic inflammation, and organs affected by metabolic disorders. The focus is on the practical integration of the nanonetwork's core architecture—sensor, processor, actuator—with complex in vivo environments to achieve targeted diagnostic and therapeutic outcomes.
Core Architecture Integration with Pathophysiology
The basic alarm-system nanonetwork comprises three functional units: 1) Biomarker Sensor (e.g., antibody, aptamer, molecularly imprinted polymer), 2) Signal Processor/Transducer (e.g., logic-gated nanoparticle, enzyme-based amplification), and 3) Effector/Actuator (e.g., drug release module, reporter signal generator). Successful deployment requires tailoring the physicochemical properties and operational logic of each unit to the unique vascular, interstitial, and cellular biology of the target pathology.
Table 1: Pathophysiological Hallmarks and Corresponding Nanonetwork Design Parameters
| Deployment Scenario | Key Biomarkers (Examples) | Physiological Barriers | Nanonetwork Design Adaptation | Primary Actuation Output |
|---|---|---|---|---|
| Solid Tumors (e.g., Breast, Pancreatic) | MMP-9, CA-IX, EGFR, Extracellular pH (~6.5-7.0) | Enhanced Permeability & Retention (EPR), High Interstitial Pressure, Dense Stroma | Size: 20-100 nm for EPR; pH- or enzyme-responsive shell; Hypoxia-sensitive logic gate. | Triggered cytotoxic release (Doxorubicin, SN-38), PDT activation. |
| Inflammation (e.g., Rheumatoid Arthritis, Colitis) | TNF-α, IL-6, ROS, Myeloperoxidase, Selectins | Inflammatory Vasodilation, Cellular Infiltrate, Reactive Oxygen Species | Size: < 150 nm; ROS-cleavable linkers; Vascular targeting ligands (e.g., anti-ICAM-1). | Release of anti-inflammatory (Dexamethasone, Tocilizumab), ROS scavenging. |
| Metabolic Disorders (e.g., NAFLD, Atherosclerosis) | ALT/AST (liver), Oxidized LDL, Caspase-3 (apoptosis), Glucose/Insulin | Endothelial Dysfunction, Steatosis/Fibrosis, Stable Plaques | Liver-targeting ligands (GalNAc); Apoptosis sensor (Annexin V logic); Enzyme-substrate probes. | Release of anti-fibrotic (Pirfenidone), Cholesterol efflux promoters, Insulin sensitizer release. |
In VivoDeployment Protocols
Deployment for Solid Tumor Targeting
This protocol details the use of an enzyme-responsive nanonetwork for targeted drug delivery to a murine xenograft model.
Experimental Protocol: MMP-9 Responsive Nanonetwork in a 4T1 Breast Cancer Model
- Nanonetwork Synthesis: Prepare PEGylated liposomal nanoparticles (100 nm) loaded with a fluorescent dye (DiR, for tracking) and doxorubicin (Dox). Functionalize the surface with a MMP-9 cleavable peptide (e.g., GPLGV) linked to a PEG corona that shields an internal cell-penetrating peptide (CPP).
- Animal Model: Inject 4T1-luc cells (1x10^6) subcutaneously into the flank of BALB/c mice. Allow tumors to grow to ~100 mm³.
- Administration & Biodistribution: Inject nanoparticles (5 mg/kg Dox equivalent) intravenously via the tail vein. At 0, 4, 24, and 48 hours post-injection, image mice using an IVIS Spectrum system (Ex/Em: 745/800 nm for DiR).
- Activation Assessment: Sacrifice animals at 48 hours. Harvest tumors and major organs. Homogenize tissues and quantify Dox fluorescence (Ex/Em: 480/590 nm) or analyze by HPLC-MS/MS. Perform immunohistochemistry for MMP-9 expression and cleaved peptide fragments.
- Efficacy Evaluation: In a separate longitudinal study, administer nanoparticles or controls (saline, free Dox) twice weekly for two weeks. Measure tumor volume daily and monitor mouse weight. At endpoint, analyze tumors for apoptosis (TUNEL assay).
Diagram Title: Nanonetwork Activation in Tumor Microenvironment
Deployment for Inflammatory Disease Targeting
This protocol outlines the use of a reactive oxygen species (ROS)-sensitive nanonetwork for targeted delivery to an inflamed joint.
Experimental Protocol: ROS-Responsive Nanonetwork in a Murine CIA Model
- Nanonetwork Synthesis: Synthesize nanoparticles from a thioketal (TK) polymer, which degrades under high ROS (H₂O₂, OH•). Load nanoparticles with dexamethasone phosphate (Dex-P) and a NIR dye (Cy7).
- Disease Model: Induce collagen-induced arthritis (CIA) in DBA/1 mice via intradermal injection of bovine type II collagen emulsified in CFA. Monitor for clinical arthritis scores.
- Biodistribution & Specificity: Upon onset of clinical arthritis (score ≥ 2), inject TK nanoparticles intravenously. Perform in vivo optical imaging at 1, 6, and 24 hours. Compare signal intensity in inflamed vs. contralateral non-inflamed paws.
- Pharmacodynamic Analysis: Extract synovial tissue 24 hours post-injection. Analyze for nanoparticle presence (fluorescence microscopy) and quantify local Dex concentration (LC-MS). Perform flow cytometry on synovial cell suspensions to assess changes in immune cell populations (e.g., decreased Ly6G+ neutrophils, CD11c+ macrophages).
- Therapeutic Study: Treat mice with 3 doses of TK-NP-Dex, free Dex, or saline every 48 hours. Monitor arthritis score, paw thickness, and perform micro-CT at endpoint to assess bone erosion.
Deployment for Metabolic Disorder Targeting
This protocol describes a two-step amplification nanonetwork for detecting and responding to hepatocyte apoptosis in non-alcoholic steatohepatitis (NASH).
Experimental Protocol: Apoptosis-Sensing Nanonetwork in a NASH Mouse Model
- Nanonetwork Design: Construct a system with a primary nanoparticle that exposes phosphatidylserine (PS) upon caspase-3/7 activation. A secondary nanoparticle containing a drug payload (e.g., caspase inhibitor or anti-fibrotic) is conjugated to an annexin V protein, which binds specifically to exposed PS.
- Animal Model: Feed C57BL/6 mice a methionine-choline deficient (MCD) diet or a high-fat, fructose, and cholesterol (FFC) diet for 8-12 weeks to induce NASH with apoptosis.
- In Vivo Activation Confirmation: Inject the primary "sensor" nanoparticle (caspase-activatable) intravenously. After 4 hours, inject the secondary "effector" nanoparticle. Harvest liver 2 hours later. Analyze liver homogenates via fluorescence resonance energy transfer (FRET) assay for caspase activity and quantify secondary NP accumulation via its tag.
- Target Engagement Verification: Perform immunofluorescence co-localization analysis on liver sections for TUNEL (apoptosis), nanoparticle signal, and α-SMA (fibrosis).
- Functional Outcome: In a therapeutic study, administer the complete two-component system 3 times weekly for 4 weeks. Assess endpoints: serum ALT/AST, liver triglyceride content, and histopathological scoring (NAS score).
Diagram Title: Two-Step Nanonetwork Logic for NASH
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for In Vivo Nanonetwork Deployment Research
| Reagent / Material | Function in Deployment Research | Example Product / Specification |
|---|---|---|
| MMP-9 Cleavable Peptide | Provides disease-specific responsiveness for tumor-targeting nanonetworks. Sensitive linker between stealth layer and active ligand. | Sequence: GPLGVRGK (custom synthesis, >95% HPLC purity). |
| Thioketal (TK) Polymer | Core material for ROS-responsive nanoparticles. Degrades selectively in inflammatory environments, enabling triggered release. | Poly(1,4-phenylenacetone dimethylene thioketal) (PPADT), Mw ~10-20 kDa. |
| Caspase-3/7 Substrate Peptide | Core sensing element for apoptosis-detecting nanonetworks. Incorporated into nanoparticles to create "always-on" to "off" fluorescence switches or masking groups. | DEVD peptide linked to a quencher/fluorophore pair (e.g., DEVDK-FITC). |
| GalNAc (N-Acetylgalactosamine) Ligand | Enables hepatocyte-specific targeting via asialoglycoprotein receptor (ASGPR) binding. Critical for liver disorder deployment. | Tris-GalNAc cluster, PEG spacer, terminal maleimide for conjugation. |
| Near-Infrared (NIR) Fluorophores | Allows real-time, non-invasive tracking of nanonetwork biodistribution and accumulation in vivo. Minimizes tissue autofluorescence. | DiR (1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindotricarbocyanine Iodide), Cy7.5 NHS ester. |
| Immunocompetent Disease Models | Provide the complete pathophysiological context (immune cells, cytokines, stroma) for testing nanonetwork performance. | Mice: ApcMin/+ (intestinal neoplasia), db/db or ob/ob (metabolic), CIA or IMQ-induced (inflammatory). |
Table 3: Quantitative Performance Metrics Across Deployment Scenarios
| Scenario | Typical NP Size (nm) | Circulation t½ (hr) | Target Accumulation (%ID/g)* | Detection Sensitivity (Biomarker Conc.) | Therapeutic Index (vs. Free Drug) |
|---|---|---|---|---|---|
| Tumor (EPR-based) | 70 - 120 | 8 - 15 | 3 - 8% ID/g | MMP-9: >10 nM; pH drop: <6.8 | 2-5x improvement |
| Inflammation (Vascular Targeting) | 100 - 150 | 4 - 10 | 5 - 12% ID/g | ROS: 50-100 µM H₂O₂ equiv. | 3-8x improvement (local toxicity) |
| Metabolic (Ligand-Directed) | 20 - 50 | 1 - 6 | 15 - 25% ID/g (liver) | Caspase-3: ~200 nM activity | 1.5-3x improvement (hepatic specificity) |
%ID/g: Percentage of injected dose per gram of target tissue.
The in vivo deployment of alarm-system nanonetworks demands a precise alignment of their architectural principles with the pathophysiological reality of the target disease. As demonstrated in tumors, inflammatory sites, and metabolically stressed tissues, success hinges on the rational selection of biomarker sensors, the engineering of robust barrier-penetrating formulations, and the logical programming of activation thresholds and effector outputs. The provided protocols and toolkits serve as a foundational guide for researchers aiming to translate nanonetwork blueprints into functionally validated, disease-responsive systems. Future advances will involve increasing network complexity (multi-input logic), integrating real-time reporting, and enhancing actuation precision to move beyond simple drug release to adaptive, closed-loop therapeutic interventions.
Navigating Nanoscale Challenges: Signal Noise, Biocompatibility, and System Optimization
Mitigating Background Noise and False Positives in Complex Biological Matrices
Within the architecture of an alarm-system nanonetwork for biomarkers research, the reliable detection of low-abundance targets amidst complex biological matrices (e.g., blood, serum, tissue lysates) is paramount. These matrices are replete with interferents—proteins, lipids, salts, and cellular debris—that contribute to significant background noise and generate false-positive signals. This technical guide details advanced strategies to enhance signal fidelity, a critical determinant for the successful translation of diagnostic and therapeutic nanonetworks.
Key sources compromising specificity in complex matrices include:
- Non-specific Adsorption (NSA): Biomolecules adhere to sensor surfaces or nanoparticle interfaces, creating a background signal.
- Matrix Effects: Viscosity, pH, ionic strength, and endogenous enzymes can alter assay kinetics and quench or amplify signals.
- Cross-reactivity: Recognition elements (e.g., antibodies, aptamers) binding to structurally similar, non-target analytes.
- Autofluorescence: Intrinsic fluorescence of matrix components in optical-based systems.
Core Mitigation Strategies: Experimental Protocols
Surface Passivation and Blocking Protocols
Objective: Minimize non-specific adsorption to sensor/nanoparticle surfaces. Detailed Protocol:
- Surface Preparation: Clean substrate (e.g., gold SPR chip, glass slide, polymer nanoparticle) via oxygen plasma treatment or piranha solution (Caution: Highly corrosive).
- Functionalization: Immerse in 2 mM solution of heterobifunctional PEG (e.g., SH-PEG-COOH) for 12 hours at 4°C to form a dense, hydrophilic, anti-fouling monolayer.
- Blocking: Incubate with a blocking buffer for 2 hours at 37°C. Composition must be empirically optimized for the matrix.
- Validation: Expose the passivated surface to the biological matrix spiked with a non-target protein (e.g., 1 mg/mL BSA in 10% serum). Measure adsorbed mass (e.g., via QCM-D). A successful passivation reduces non-specific adsorption by >90%.
Table 1: Efficacy of Common Blocking Agents in Human Serum
| Blocking Agent | Concentration | Application | % Reduction in NSA (vs. unblocked) |
|---|---|---|---|
| Bovine Serum Albumin (BSA) | 1-5% w/v | General purpose, immunoassays | 70-85% |
| Casein | 1-2% w/v | Phosphoprotein studies, ELISA | 75-88% |
| Fish Skin Gelatin | 0.1-1% | High sensitivity assays | 80-90% |
| PEG-based Blockers | 1% | Nanoparticle functionalization | 90-95% |
| Commercial Blocker | As per mfr. | Challenging matrices (e.g., lysate) | 85-92% |
Affinity Probe Engineering for Enhanced Specificity
Objective: Increase binding affinity and specificity of recognition elements. Detailed Protocol:
- Aptamer Selection (SELEX) with Counter-SElection:
- Perform standard SELEX against the purified target biomarker.
- Critical Step: Introduce counter-selection rounds against the full, depleted biological matrix (e.g., serum from healthy controls) before exposure to the target. This removes sequences binding to common matrix interferents.
- Use high-throughput sequencing (NGS) to identify dominant families post-selection.
- Antibody Validation via Western Blot/MS: Confirm monoclonal antibody specificity not just against recombinant protein, but via immunoprecipitation from the native matrix followed by mass spectrometry to identify all pulled-down proteins.
Signal Amplification with Background Suppression
Objective: Amplify target signal while suppressing background. Detailed Protocol: Proximity Ligation Assay (PLA) in Tissue
- Probe Binding: Apply two target-specific antibodies conjugated to unique DNA oligonucleotides to a formalin-fixed tissue section.
- Proximity Ligation: Only when both antibodies bind in close proximity (<40 nm), a connector oligonucleotide hybridizes, enabling ligation to form a closed circular DNA template.
- Rolling Circle Amplification (RCA): Add Phi29 DNA polymerase and nucleotides. Circular DNA template generates a long, repeating single-stranded DNA product only at the site of the target.
- Detection: Detect RCA product with fluorescently labeled complementary oligonucleotides. This confines massive signal amplification exclusively to genuine dual-epitope binding events, virtually eliminating false positives from single-antibody cross-reactivity.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Noise Mitigation Experiments
| Item | Function | Example Product/Catalog # |
|---|---|---|
| Heterobifunctional PEG | Creates dense, hydrophilic, anti-fouling layers on surfaces. | mPEG-SVA, 5kDa (JenKem Tech) |
| Commercial Blocking Buffer | Ready-to-use, optimized blends of proteins/polymers for specific matrices. | Blocker BLOTTO (Thermo Fisher) |
| SPR/QCM-D Sensor Chips | Gold or silica chips for real-time, label-free measurement of binding and fouling. | Gold Sensor Chips (Biacore/Cytiva) |
| Depleted/Synthetic Matrices | Matrices stripped of specific interferents (e.g., IgG, albumin) for assay development. | Human Serum, Charcoal Stripped (Sigma) |
| Cross-reactive Adsorbent | Removes interfering antibodies from detection systems. | Adsorbent (Antibody Registry) |
| Phi29 DNA Polymerase | Enzyme for isothermal, high-fidelity Rolling Circle Amplification (RCA). | Phi29 DNA Pol (NEB) |
| NGS Library Prep Kit | For high-throughput sequencing of aptamer pools post-SELEX. | NextFlex (PerkinElmer) |
Quantitative Data Analysis & Validation
Table 3: Metrics for Assessing Noise Mitigation Performance
| Metric | Formula/Description | Acceptable Threshold (Diagnostic) |
|---|---|---|
| Signal-to-Background Ratio (S/B) | Mean(Signal) / Mean(Background) | > 10 |
| Limit of Detection (LoD) | 3.3 * (StdDev of Blank) / Slope of Calibration Curve | Biomarker-dependent (fM-pM) |
| % Coefficient of Variation (CV) | (StdDev / Mean) * 100 (across replicates) | Intra-assay: <10%, Inter-assay: <15% |
| Recovery in Spike-In | (Measured Conc. in Matrix / Expected Conc.) * 100 | 80-120% |
| Z'-Factor (HTS) | 1 - [ (3σpositive + 3σnegative) / |μpositive - μnegative| ] | > 0.5 (Excellent assay) |
Integrated Workflow & Pathway Diagrams
Alarm System Noise Mitigation Workflow
Blocked vs. Mitigated Signaling Pathways
Integrating rigorous surface chemistry, engineered high-fidelity probes, and background-suppressive amplification into the foundational architecture of biomarker-detecting nanonetworks is non-negotiable for achieving clinical-grade reliability. The systematic application of the protocols and validation metrics outlined herein provides a roadmap to transform a promising proof-of-concept into a robust alarm system capable of operating in the noisy reality of human biology.
Within the architecture of an alarm-system nanonetwork for biomarkers research, a critical functional layer is the interface between the synthetic nanodevice and the complex biological milieu. The primary objective is to deploy a network of sensors and communication nodes that can detect early pathological signatures in vivo without triggering immune surveillance, thereby maximizing operational longevity and signal fidelity. This necessitates sophisticated surface engineering to achieve both biocompatibility (minimal non-specific interactions and toxicity) and immune evasion (avoidance of opsonization and clearance by the mononuclear phagocyte system). This guide details the core principles, materials, and experimental protocols for functionalizing nanoparticle surfaces to create "stealth" nanonetwork components.
Core Principles of Stealth Coatings
The dominant strategy for immune evasion is the creation of a hydrophilic, neutrally-charged, and highly dynamic surface that minimizes protein adsorption (opsonization). The most established and effective method is the grafting or adsorption of poly(ethylene glycol) (PEG) and its derivatives, a process known as PEGylation. Recent advancements have expanded the toolkit to include zwitterionic polymers, polysaccharides, and biomimetic coatings.
Key Mechanisms:
- Steric Repulsion: Dense, hydrated polymer brushes generate a repulsive entropic barrier.
- Reduced Interfacial Free Energy: Hydrophilic coatings decrease the thermodynamic driving force for protein adhesion.
- Molecular Conformation: Flexible polymer chains (e.g., PEG) create a dynamic "cloud" that is difficult for opsonins to bind.
Quantitative Data: Coating Performance Metrics
The efficacy of stealth coatings is quantified through in vitro and in vivo experiments. Key performance indicators are summarized below.
Table 1: In Vitro Performance of Common Stealth Coatings
| Coating Type | Common Materials | Hydrodynamic Size Increase (nm)⁴ | Zeta Potential (mV)⁴ | Protein Adsorption Reduction (% vs. bare NP)⁴ | Primary Immune Evasion Mechanism |
|---|---|---|---|---|---|
| PEG (Linear) | mPEG-Thiol, NHS-PEG | 5 - 15 | -10 to +10 | 70 - 90% | Steric Repulsion, Hydration |
| Branched PEG | PEG-Dendrons | 10 - 25 | ~0 | 85 - 95% | Enhanced Surface Density & Conformation |
| Zwitterionic | PCBMA, PSBMA | 8 - 20 | ~0 | 90 - 98% | Super-hydrophilicity, Electrostatic Hydration |
| Polysaccharide | Hyaluronic Acid, Dextran | 10 - 30 | -15 to -30 | 60 - 85% | Steric Repulsion, Biomimicry |
| Cell Membrane | RBC Membrane Vesicles | 15 - 40 | -20 to -25 | 95%+ | "Self" Marker Display, CD47 Integration |
Table 2: In Vivo Pharmacokinetic Impact of Stealth Coatings (Mouse Model)⁴
| Coating Type | Circulation Half-life (t₁/₂, h) | Liver & Spleen Accumulation (%ID/g at 24h) | Key Clearance Pathway |
|---|---|---|---|
| Uncoated (Citrated) | 0.5 - 2 | 60 - 80% | Rapid Opsonization, MPS Uptake |
| Dense PEG Brush | 12 - 36 | 15 - 30% | Slow Mononuclear Phagocytosis |
| Zwitterionic Polymer | 20 - 48 | 10 - 25% | Minimal MPS Recognition |
| RBC Membrane Cloak | 30 - 72 | 5 - 20% | Evasion via "Self" Signature |
⁴ Representative data compiled from recent literature (2022-2024). Actual values depend on core NP size, material, grafting density, and animal model.
Experimental Protocols
Protocol: Covalent PEGylation of Gold Nanoparticles (AuNPs) via Thiol Chemistry
Objective: To create a dense monolayer of methoxy-PEG-thiol (mPEG-SH) on a 50nm spherical AuNP core.
Materials: See "The Scientist's Toolkit" below. Procedure:
- NP Activation: Concentrate 1 mL of citrate-stabilized 50nm AuNPs (OD₅₂₀ ~5) via centrifugation (10,000 x g, 15 min). Resuspend in degassed, deionized water.
- Ligand Exchange: Add a 10,000-fold molar excess of mPEG-SH (5 kDa) to the AuNP suspension. Vortex immediately.
- Reaction: Incubate the mixture at room temperature for 16-24 hours with gentle agitation, protected from light.
- Purification: Remove unreacted PEG via three cycles of centrifugation (8,000 x g, 20 min) and resuspension in 1x PBS (pH 7.4).
- Characterization:
- DLS: Confirm increase in hydrodynamic diameter and polydispersity index (PDI) < 0.2.
- Zeta Potential: Measure in 1 mM KCl; successful PEGylation shifts potential towards neutral (~0 to -5 mV).
- UV-Vis: Verify no significant plasmon band shift or broadening, indicating colloidal stability.
Protocol: Assessing Stealth Properties via Protein Corona Analysis (SDS-PAGE)
Objective: To qualitatively compare the protein adsorption profiles of coated vs. uncoated nanoparticles.
Procedure:
- Incubation with Serum: Incubate 100 µL of each NP formulation (1 mg/mL in PBS) with 100 µL of 100% fetal bovine serum (FBS) at 37°C for 1 hour.
- Corona Isolation: Separate NP-protein complexes from unbound proteins by centrifugation (conditions as in 4.1, step 4). Wash pellet gently with PBS twice.
- Protein Elution: Resuspend the final pellet in 30 µL of 2x Laemmli SDS-PAGE sample buffer containing 5% β-mercaptoethanol. Heat at 95°C for 10 minutes to denature and elute proteins.
- Analysis: Load supernatant onto a 4-20% gradient polyacrylamide gel. Run electrophoresis alongside a molecular weight marker. Stain with Coomassie Blue or silver stain.
- Interpretation: Effective stealth coatings will show fainter and fewer protein bands compared to uncoated NPs.
Diagrams
Immune Clearance of Uncoated Nanoparticles
Stealth Coating Mechanism of Action
Surface Functionalization Workflow
The Scientist's Toolkit
Table 3: Essential Reagents for Surface Functionalization & Stealth Coating Research
| Item | Function & Rationale |
|---|---|
| Gold Nanoparticles (Citrate-stabilized, 20-100nm) | A standard, well-characterized model nanoparticle core for method development due to easy surface modification and strong plasmonic signature. |
| Methoxy-PEG-Thiol (mPEG-SH, 2-10 kDa) | The benchmark "stealth" polymer. Thiol group provides strong, covalent attachment to gold and other metal surfaces. |
| Phosphorylcholine-based Zwitterionic Monomer (e.g., MPC) | For synthesizing ultra-low fouling polymer brushes via surface-initiated polymerization (e.g., ATRP). |
| DSPE-PEG(2000)-COOH / -NH₂ | Phospholipid-PEG conjugates for functionalizing lipid-based NPs (liposomes, micelles) and introducing click chemistry handles. |
| Hyaluronic Acid (Low MW, modified with ADH or NHS) | A natural polysaccharide for biomimetic coatings; can also target CD44 receptors on some cell types. |
| EZ-Link NHS-Biotin / Streptavidin | A versatile bioconjugation toolkit for attaching targeting ligands (antibodies, peptides) to pre-coated stealth nanoparticles. |
| Fetal Bovine Serum (FBS) | A complex protein mixture used for in vitro protein corona formation assays to predict in vivo behavior. |
| Dynamic Light Scattering (DLS) / Zeta Potential Analyzer | Essential instrument for measuring hydrodynamic size, polydispersity, and surface charge before/after coating. |
| SDS-PAGE Gel Electrophoresis System | For separating and visualizing proteins adsorbed from serum to form the "corona," a key metric of stealth efficacy. |
Addressing Power and Energy Harvesting Constraints for Sustained Operation
Within the thesis framework of a basic architecture for an alarm-system nanonetwork for biomarker research, sustained operation is the paramount challenge. Such a network, comprising nanoscale sensors, processors, and communicators deployed in vivo, must function autonomously for extended periods to monitor disease biomarkers. This whitepaper provides an in-depth technical analysis of contemporary power and energy harvesting constraints, presenting current solutions, experimental protocols, and material toolkits to enable long-term functionality.
Current Energy Harvesting Modalities forIn VivoNanonetworks
The operational lifetime of an implantable nanonetwork is dictated by its energy budget. The table below summarizes the performance characteristics of leading energy harvesting modalities, based on recent (2023-2024) experimental studies.
Table 1: Quantitative Comparison of In Vivo Energy Harvesting Modalities
| Modality | Power Density (Typical) | Voltage Output | Key Advantage | Primary Constraint |
|---|---|---|---|---|
| Biochemical (Glucose/O₂ Fuel Cell) | 10–100 µW/cm² | 0.2–0.8 V | Utilizes abundant physiological fuels (glucose, O₂). | Low power density; enzyme/biocatalyst stability. |
| Piezoelectric (Mechanical Motion) | 10–300 µW/cm³ | 1–5 V (AC) | High voltage from continuous physiological motion (heartbeat, breathing). | Inconsistent/irregular input; requires rectification. |
| Triboelectric Nanogenerators (TENGs) | 1–50 mW/cm³ (peak) | 10–100 V (AC) | Exceptionally high peak power from interfacial contact. | High impedance, voltage management, long-term material wear. |
| Radiofrequency (RF) Harvesting | 0.1–10 µW/cm² (at depth) | 0.5–3 V | External power can be controlled on-demand. | Rapid attenuation in tissue; strict regulations on transmit power. |
| Photovoltaic (Subdermal) | 10–50 µW/cm² (under skin) | 0.5–1.0 V | Stable output if sufficiently illuminated. | Limited light penetration through tissue (< 10 mm). |
Core Architecture: Integrated Energy Management Unit (EMU)
For sustained operation, the nanonetwork node must integrate an Energy Management Unit (EMU) that orchestrates harvesting, storage, and consumption. The logical workflow is defined below.
Diagram Title: Energy Management Unit (EMU) Data & Power Flow
Experimental Protocol: Evaluating Piezoelectric Harvesters for Vascular Implants
This protocol details a method to characterize a flexible piezoelectric energy harvester's performance under simulated physiological pulsation.
Title: In Vitro Characterization of a PZT-PDMS Composite Harvester.
Objective: To measure the voltage, current, and sustained power output of a nanogenerator under cyclic mechanical strain mimicking arterial pulse.
Materials: See "Research Reagent Solutions" (Section 6).
Methodology:
- Device Fabrication: Spin-coat a 100 µm thick PDMS layer on a flexible substrate. Deposit interdigitated electrodes (IDE) via sputtering. Doctor-blade a composite paste of PZT nanoparticles (70% wt) in PDMS matrix onto the IDE. Cure at 80°C for 2 hours.
- Test Setup: Mount the fabricated harvester on a programmable pneumatic actuator. Connect the harvester's electrodes to a high-input-impedance oscilloscope (for voltage) and a source measure unit (SMU) for current/power.
- Simulated Pulse Profile: Program the actuator to apply a sinusoidal strain of 5-10% at a frequency of 1.2 Hz (72 beats/minute) with a 30% duty cycle.
- Data Acquisition:
- Record the open-circuit voltage (Voc) and short-circuit current (Isc) over 10,000 cycles.
- Perform a load sweep using the SMU. Connect variable load resistors (1 kΩ to 10 MΩ) across the harvester output. Measure the RMS voltage (Vrms) across each load.
- Calculate power delivered: Pload = (Vrms)² / Rload.
- Identify the optimal load resistance (Ropt) for maximum power transfer (Pmax).
- Endurance Testing: Subject the harvester to 1 million cycles. Record Voc and Pmax at intervals of 100k cycles to assess performance degradation.
Signaling Pathway for Activity-Driven Duty Cycling
To minimize energy consumption, the nanonetwork's operational state must be governed by biomarker detection events. The following logic pathway enables ultra-low-power standby with event-triggered activation.
Diagram Title: Biomarker-Triggered Activation Logic
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Nanonetwork Energy Harvester Fabrication & Testing
| Item | Function/Description | Example Product/Specification |
|---|---|---|
| PZT Nanoparticles | High piezoelectric coefficient material for mechanical energy conversion. | Lead zirconate titanate, ~100 nm diameter, 99% purity. |
| Polydimethylsiloxane (PDMS) | Flexible, biocompatible polymer matrix for composite harvesters. | Sylgard 184 Silicone Elastomer Kit. |
| Interdigitated Electrode (IDE) Mold | Creates efficient charge collection geometry on flexible substrates. | Photolithographically patterned Si wafer with ~20 µm line spacing. |
| Programmable Biomechanical Actuator | Simulates physiological motion (pulse, respiration) for in vitro testing. | Bose ElectroForce 3100 with custom soft grip fixtures. |
| Source Measure Unit (SMU) | Precisely sweeps load resistance and measures µW-level power output. | Keithley 2450 SMU with low-current sensitivity. |
| Flexible Supercapacitor | Micro-energy storage component for power buffering. | Graphene-based, solid-state, >100 µF/cm² areal capacitance. |
| Biochemical Fuel Cell Enzymes | Biocatalysts for glucose/O² oxidation/reduction in physiological fuel cells. | Glucose oxidase (GOx) and Laccase, immobilized on CNT electrodes. |
| RFID/NFC Reader & Harvesting IC | For testing RF energy harvesting link budget and power conversion. | Texas Instruments RF430FRL152H, includes integrated harvester. |
Optimizing Network Density and Topology for Reliable Coverage and Robustness
The basic architecture of an alarm-system nanonetwork is predicated on a distributed, node-based system of nanoscale sensors designed for in vivo biomarker detection. This network's primary function is to detect specific molecular signatures (e.g., cytokines, cell-free DNA, exosomes) and transmit a coordinated signal to an external receiver, triggering an alert for therapeutic intervention. Within this thesis, optimizing network density (number of nodes per unit volume) and topology (physical/spatial arrangement and communication links) is critical for ensuring complete coverage of a target tissue and robustness against node failure or dynamic biological clearance.
Core Principles: Density, Topology, and Performance Metrics
Performance is evaluated against three core metrics:
- Coverage: The percentage of the target volume within sensing range of at least one nanodevice.
- Robustness: The network's ability to maintain connectivity and signal fidelity despite node degradation, movement, or loss.
- Energy/Lifetime Efficiency: Minimizing inter-node communication load to prolong in vivo operational duration.
These metrics are inherently in tension. High random density increases coverage but can cause interference and rapid resource depletion. A structured topology can enhance robustness but may be difficult to achieve in vivo. Optimization finds the Pareto-optimal balance.
Quantitative Analysis of Topology-Density Trade-offs
Recent simulation and in vitro experimental studies provide the following comparative data.
Table 1: Performance of Common Nanonetwork Topologies vs. Density
| Topology Type | Optimal Node Density (nodes/mm³) | Achievable Coverage (%) | Path Redundancy (Avg. # of Paths) | Estimated Robustness to 20% Node Loss |
|---|---|---|---|---|
| Random Uniform | 50 - 100 | 70 - 85 | Low (1.2) | Poor (<50% coverage retained) |
| Regular Grid (Lattice) | 20 - 30 | 90 - 95 | Medium (2.0) | Medium (~70% coverage retained) |
| Scale-Free (Hub-Based) | 10 - 15 | 60 - 75 | High (3.5) | High (if hubs survive) / Very Low (if hubs fail) |
| Small-World (Clustered) | 30 - 50 | 95 - 98 | Medium-High (2.8) | High (~85% coverage retained) |
| Gradient (Density-tapered) | 40 (core) - 10 (edge) | 92 - 96 | Medium (2.3) | Medium-High (~80% coverage retained) |
Table 2: Signaling Modalities and Their Network Implications
| Signaling Modality | Max Range | Bandwidth | Energy Cost per Bit | Suitability for Dense Networks |
|---|---|---|---|---|
| Molecular Diffusion | µm-scale | Very Low | High (chem. synthesis) | Poor - High interference in density |
| Acoustic / Ultrasonic | cm-scale | Medium | Low | Excellent - Low cross-talk |
| Magnetic Resonance | mm-cm scale | Low | Medium | Good - Addressable nodes |
| Near-Infrared (NIR) FRET | nm-µm scale | High | Medium | Fair - Requires very close proximity |
| Radiofrequency (EM) | mm-scale | Very Low | Very High | Poor in tissue (attenuation) |
Experimental Protocols forIn VitroValidation
Protocol 4.1: Microfluidic Chamber for Coverage and Connectivity Mapping
Objective: To empirically determine the relationship between injected node density and coverage of a 2D surface simulating tissue. Materials:
- PDMS microfluidic chamber with a 10mm x 10mm x 0.05mm main channel.
- Fluorescently-labeled nanoparticle analogs (e.g., 200nm liposomes with surface-conjugated "sensor" antibodies).
- Buffer solution (PBS, pH 7.4).
- Programmable syringe pump.
- Confocal fluorescence microscope with Z-stack capability.
- Image analysis software (e.g., Fiji/ImageJ with custom macros). Methodology:
- Prime the chamber with buffer.
- Infuse nanoparticle suspension at a calculated concentration (e.g., 10, 50, 100 particles/nL) using the syringe pump at 1 µL/min.
- Allow particles to settle/adhere for 30 minutes under static conditions.
- Gently perfuse buffer to remove non-adhered particles.
- Acquire tiled confocal Z-stack images of the entire chamber bottom.
- Process images: threshold fluorescence, identify particle centroids, and calculate Voronoi tessellation.
- Coverage = (Total area of Voronoi cells with radius ≤ sensing range) / (Total chamber area).
- Connectivity: Apply a distance threshold (max communication range) to particle centroids to generate an adjacency matrix. Analyze the graph for connected components and path redundancy.
Protocol 4.2: Robustness Assay via Enzymatic Node Degradation
Objective: To test network signal propagation fidelity under simulated node failure. Materials:
- Network of DNA-origami nodes with enzymatic cleavage sites and FRET-based signaling.
- Target-specific protease (e.g., MMP-9, simulating a biomarker-triggered and a background enzyme).
- Fluorescence plate reader or real-time PCR system (for fluorescence kinetics).
- Control buffer. Methodology:
- Assemble a small-world network of DNA-origami nodes in a well plate. Establish a baseline signal cascade from a designated "source" to "reporter" node.
- Treat the network with a low concentration of protease (e.g., 0.1 U/mL MMP-9).
- Monitor the fluorescence output of the reporter node in real-time over 60 minutes.
- Repeat the cascade initiation protocol every 10 minutes.
- Quantify the Signal Retention Index (SRI) = (Peak Fluorescence at Tn / Peak Fluorescence at T0) * 100%.
- Correlate SRI decay rate with the initial network topology, as verified by atomic force microscopy (AFM) at experiment start and end.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Nanonetwork Research
| Item / Reagent | Function in Network Research | Example Product/Chemical |
|---|---|---|
| Functionalized Liposomes (200nm) | Prototype nanonode; carrier for sensors/transmitters. | Avanti Polar Lipids, DSPC/Cholesterol PEGylated. |
| DNA Origami Kit | For constructing precise, programmable topological structures. | Tilibit Nanosystems "M13mp18" Scaffold Kit. |
| Heterobifunctional PEG Linkers | Conjugation of ligands, antibodies, or signaling molecules to node surface. | Thermo Fisher Scientific, SM(PEG)n reagents. |
| FRET Pair Donor/Acceptor Dyes | Enables proximity-based inter-node communication. | Cy3/Cy5 (Donor/Acceptor) from Lumiprobe. |
| Microfluidic Chamber (PDMS) | Provides a controlled 3D environment for network assembly and testing. | Synder µSil or custom fabricated via soft lithography. |
| Protease (MMP-9) | Simulates dynamic in vivo degradation of protein-based node components. | R&D Systems, recombinant human MMP-9. |
| Quartz Crystal Microbalance with Dissipation (QCM-D) | Measures real-time adsorption and interaction of nodes on surfaces. | Biolin Scientific QSense Analyzer. |
Signaling Pathway & System Workflow Visualizations
Alarm Nanonetwork Signaling Cascade
Scale-Free with Small-World Clusters Topology
Nanonetwork Optimization Workflow
Abstract This whitepaper details the security architecture for an alarm-system nanonetwork designed for biomarker research, a critical subsystem within the broader thesis on the Basic Architecture of an Alarm-System Nanonetwork for Biomarkers Research. As nanoscale devices move from sensing to actuation in therapeutic applications, ensuring the integrity of transmitted signals against spoofing and interference becomes paramount. We present a technical guide for implementing cryptographic and physical-layer protocols to authenticate commands and data within a body area nanonetwork.
1. Introduction: The Spoofing Threat in Therapeutic Nanonetworks An alarm-system nanonetwork comprises three core components: 1) Sensor Nodes: Engineered nanoparticles or synthetic cells that detect specific biochemical biomarkers; 2) Relay/Amplifier Nodes: Intermediate nanodevices that propagate signals; 3) Actuator Nodes: Nanomachines that release drugs or perform corrective actions upon authenticated alarm signals. A spoofed signal—an illegitimate signal mimicking a valid alarm—could trigger premature, excessive, or toxic therapeutic responses. Securing this network requires solutions adaptable to severe constraints in computational power, energy, and bandwidth at the nanoscale.
2. Core Security Paradigms and Quantitative Analysis We evaluate two primary, non-mutually exclusive paradigms for nanoscale signal authentication: Physical-Layer Security and Lightweight Cryptography.
Table 1: Comparison of Nanoscale Security Paradigms
| Paradigm | Core Principle | Energy/Compute Footprint | Spoofing Resistance | Best Application Context |
|---|---|---|---|---|
| Physical-Layer Fingerprinting | Exploits unique, hard-to-clone physical characteristics of the communication channel or device. | Very Low | High vs. external spoofers; Medium vs. compromised internal nodes. | Closed, homogeneous nanonetworks with stable environmental conditions. |
| Molecular Barcoding / Timing | Embeds authentication within the physical properties of the signaling molecule (e.g., isotopic ratio) or the temporal pattern of release. | Low (for sensing) | High vs. all spoofers, provided the barcode secret is not leaked. | Molecular communication (MC)-based nanonetworks. |
| Lightweight Symmetric Cryptography | Uses pre-shared secret keys and minimalistic algorithms (e.g., PRESENT, SPONGENT) to generate message authentication codes (MACs). | Medium (requires nano-processor design) | Very High, conditional on key secrecy and storage integrity. | Digital electromagnetic or acoustic nanonetworks with integrated circuitry. |
3. Experimental Protocols for Security Validation
Protocol 3.1: Validating Molecular Barcode Authentication Objective: To confirm that a spoofed signal using natural abundance ligands does not trigger an actuator node tuned to a specific isotopic barcode. Materials: See Scientist's Toolkit. Procedure:
- Synthesis: Prepare two batches of the same signaling molecule (e.g., a peptide). Batch A: synthesized with (^{12})C and (^{14})N atoms (natural abundance). Batch B: synthesized with >99% (^{13})C at three specific backbone positions.
- Functionalization: Functionalize gold nanoparticle (Actuator Node) surfaces with receptors specifically engineered for high-affinity binding to the isotopically heavy motif of Batch B, leveraging kinetic isotopic effects.
- Spoofing Simulation: Introduce Batch A (spoof signal) into the experimental microfluidic chamber at 10x the anticipated physiological concentration of the target biomarker. Monitor actuator node (e.g., drug release) via fluorescence de-quenching.
- Genuine Alarm Test: Introduce Batch B (authentic signal) at the minimum biomarker threshold concentration. Monitor activation.
- Control: Co-incubate Batch B with a 100-fold excess of Batch A to test specificity under interference. Data Analysis: Activation should be statistically significant (p < 0.01) only for steps 4 & 5, confirming spoofing resistance.
Protocol 3.2: Testing Physical-Layer RF Fingerprinting Objective: To differentiate between legitimate intra-body nanotransmitters and external spoofers using RF distinct native attributes. Materials: Custom nanoscale RF emitters, Software-Defined Radio (SDR) receiver, CNN classifier. Procedure:
- Legitimate Signal Capture: Power a population of 100 identical nanotransmitters sequentially in a saline bath. Capture 10,000 transient RF bursts per device using the SDR. Extract features (I/Q imbalance, turn-on transient, spectral mask).
- Spoofer Signal Capture: Use a macroscopic lab emitter to mimic the nominal frequency and modulation of the nanotransmitters. Capture equivalent data.
- Model Training: Train a convolutional neural network (CNN) on 80% of the data to classify "Legitimate Node 1...100" vs. "External Spoofer."
- Validation: Test on the held-out 20%. The confusion matrix must show >99.5% accuracy in identifying the external spoofer class. Data Analysis: Successful deployment requires the classifier model to be embedded in the network's base station (e.g., a wearable hub).
4. Integrated Security Architecture for the Alarm-System Nanonetwork The proposed architecture implements defense in depth, combining the above paradigms based on communication modality.
Table 2: Layered Security Implementation
| Network Layer | Communication Modality | Primary Security Mechanism | Fallback Mechanism |
|---|---|---|---|
| Sensor → Relay | Molecular Diffusion / Quorum Sensing | Molecular Barcoding (Isotopic) | Pre-shared Concentration Threshold & Temporal Pattern Lock |
| Relay → Relay | Acoustic / Piezoelectric | Physical-Layer Fingerprinting (Transient Response) | -- |
| Relay → Actuator | Targeted Molecular Delivery / RF | Lightweight MAC (PRESENT-80) | Physical-Layer Fingerprinting |
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Nanoscale Security Experiments
| Item | Function | Example Product/Catalog # |
|---|---|---|
| Isotopically Labeled Amino Acids | Enables synthesis of molecular barcodes for authentication. | (^{13})C(_6)-L-Lysine (Cambridge Isotope, CLM-226) |
| Functionalized Gold Nanoparticles (10-50nm) | Platform for constructing actuator nodes with surface-grafted cryptographic receptors. | Citrate-coated AuNPs, 30nm (Sigma-Aldrich, 753610) |
| Microfluidic Organ-on-a-Chip | Provides a physiologically relevant environment for testing spoofing in simulated tissue. | Emulate, Inc. Liver-Chip or similar. |
| Software-Defined Radio (SDR) Platform | For capturing and analyzing physical-layer RF characteristics of nanodevices. | Ettus Research USRP B210 |
| Lightweight Crypto IP Core | FPGA or ASIC design for implementing algorithms like PRESENT for MAC generation. | OpenCores.org PRESENT cipher core |
5. Visualizing Signaling Pathways and Security Workflows
Diagram 1: Alarm System Data Flow & Spoofing Block
Diagram 2: Security Mechanism Decision Workflow
6. Conclusion and Future Directions Securing alarm-system nanonetworks demands a multi-layered approach tailored to nanoscale constraints. Molecular barcoding and physical-layer fingerprinting offer promising low-energy solutions, while minimalist cryptography provides robust authentication where minimal computation is feasible. Future research must focus on in vivo validation of these protocols and the development of unified security standards for nanomedical devices, ensuring that therapeutic actions are triggered only by authentic biological alarms.
Benchmarking Performance: Validation Frameworks and Comparative Technology Analysis
The development of a basic architecture for an alarm-system nanonetwork for biomarkers research necessitates a foundational, predictive framework. This architecture envisions deployable nanoscale devices capable of detecting, processing, and communicating specific biomarker signals in vivo. Prior to costly physical fabrication and biological validation, in silico modeling and simulation platforms serve as the indispensable gold standard for de-risking design, optimizing communication protocols, and predicting system behavior in complex biological environments.
Core Platform Architectures and Quantitative Comparison
Current in silico platforms can be categorized by their modeling approach, each offering distinct advantages for simulating nanonetwork components.
Table 1: Comparison of Primary In Silico Modeling Platforms for Nanonetwork Research
| Platform Name / Type | Primary Modeling Approach | Key Strengths for Alarm-System Nanonetworks | Computational Demand | Example Tools / Libraries |
|---|---|---|---|---|
| Molecular Dynamics (MD) | Atomistic/Physics-based | High-fidelity ligand-receptor binding, nanoparticle diffusion, molecular conformation. | Extremely High | GROMACS, NAMD, AMBER |
| Stochastic Simulation (SSA) | Probabilistic/Chemical Master Equation | Accurate for low-copy number biochemical reactions (e.g., biomarker capture, signal transduction within a node). | Moderate to High | COPASI, StochPy, BioSimulator.jl |
| Agent-Based Modeling (ABM) | Discrete-Event/Rule-based | Ideal for individual nanomachine behavior, node-to-node communication, and emergent network dynamics. | Variable (Scales with agent count) | NetLogo, MASON, PhysiCell |
| Multi-Scale Hybrid | Integrative (Combines above) | Links molecular events to network-level outcomes; essential for full-system simulation. | Very High | Custom frameworks (e.g., coupling LAMMPS with NS-3) |
Detailed Experimental Protocols forIn SilicoValidation
Protocol 3.1: Simulating Biomarker Capture and Initial Signal Generation
Objective: To model the binding kinetics of a target biomarker to a functionalized nanosensor surface and the subsequent generation of an internal chemical signal.
- System Definition: Using a tool like COPASI, define the chemical species: Free Biomarker
[B], Free Receptor[R]on the nanodevice, Biomarker-Receptor Complex[BR], and Internal Signal Molecule[S]. - Reaction Schema: Implement the following reactions with appropriate kinetic rates (e.g., from literature or MD simulations):
B + R <-> BR(Forward ratek_on, Reverse ratek_off)BR -> R + S(Catalytic ratek_cat)
- Parameterization: Set initial concentrations:
[B]_0as estimated local biomarker concentration (e.g., 1 pM to 1 nM),[R]_0based on device surface area. Setk_on,k_offfrom surface plasmon resonance data,k_catfrom enzyme kinetics. - Simulation Execution: Run a stochastic simulation (Gibson & Bruck algorithm) for a simulated time equal to the desired detection window (e.g., 1000 seconds).
- Output Analysis: Plot
[S](t)over time. Calculate the time-to-detection threshold and signal-to-noise ratio based on basal[S]production.
Protocol 3.2: Simulating Diffusive Molecular Communication between Nanonodes
Objective: To model the propagation of a messenger molecule (e.g., Ca²⁺, IP₃, synthetic particle) from a transmitting to a receiving nanonode.
- Environment Setup: In an ABM platform like NetLogo, define a 2D or 3D grid representing the tissue microenvironment. Set diffusion coefficients for the messenger molecule (D ~ 10⁻¹⁰ to 10⁻¹² m²/s in cytoplasm).
- Node Deployment: Place transmitting and receiving agent-nodes at a fixed distance (e.g., 1-10 µm). The transmitter agent is activated based on results from Protocol 3.1.
- Communication Rule: Upon activation, the transmitter releases
Nmolecules (e.g., 1000-10000) at its location. - Diffusion Model: Implement Brownian motion for each molecule:
Δx = sqrt(2*D*Δt) * random_normal()for each time stepΔt. - Reception Logic: The receiving node counts molecules within its physical boundary per time step. A reception event is logged when the count exceeds a defined threshold.
- Metrics: Calculate and plot the hit count over time at the receiver. Determine the bit error rate for repeated pulses under varying diffusion noise.
Visualization of Key Signaling and Workflow Pathways
Diagram 1: Molecular signaling pathway within a nanonetwork node.
Diagram 2: Iterative in silico workflow for nanonetwork design.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential "Reagents" for In Silico Nanonetwork Experiments
| Item / Software | Category | Function in the In Silico Experiment |
|---|---|---|
| GROMACS | Molecular Dynamics Engine | Simulates atomic-level interactions between biomarkers, sensor surfaces, and the solvent to derive binding kinetics and diffusion coefficients. |
| COPASI | Biochemical Network Simulator | Solves systems of biochemical reactions using deterministic or stochastic algorithms to model signal generation and amplification inside a nanodevice. |
| NetLogo | Agent-Based Modeling Environment | Provides a programmable platform to simulate the behavior and interactions of thousands of individual nanomachines in a spatial environment. |
| Python (SciPy/NumPy) | General-Purpose Programming | The foundational "buffer" for custom scripting, data analysis, visualization, and coupling different simulation tools into a hybrid workflow. |
| Protein Data Bank (PDB) File | Molecular Structure Data | Provides the 3D atomic coordinates of biomarkers (e.g., proteins) or receptors necessary for initiating MD simulations. |
| Experimental Kinetic Data (e.g., from BRENDA) | Kinetic Parameter Database | Supplies critical real-world parameters (Km, Kcat, Ki) to parameterize in silico models, grounding them in biological reality. |
| High-Performance Computing (HPC) Cluster | Computational Infrastructure | Provides the necessary processing power and memory to run MD, large-scale ABM, or hybrid simulations within a feasible timeframe. |
This document details the critical validation phase for the Basic Architecture of an Alarm-System Nanonetwork for Biomarkers Research. This nanonetwork consists of engineered biosensors, signal transducers, and communication modules designed to detect specific biomarkers in vivo. The ultimate goal is to generate a quantifiable network output (e.g., fluorescence intensity, radio signal, secreted reporter) that correlates precisely with pathophysiological states. Validation across ex vivo and animal models is essential to confirm diagnostic sensitivity, specificity, and predictive value before clinical translation.
Ex VivoValidation: Precision in a Controlled System
Core Experimental Protocol
- Objective: To establish a baseline correlation between nanonetwork output and biomarker concentration in a physiologically relevant matrix.
- Methodology:
- Sample Preparation: Human or animal-derived tissues/fluids (e.g., diseased vs. healthy plasma, tumor homogenates) are collected and spiked with a gradient of the target biomarker.
- Nanonetwork Incubation: A standardized dose of the alarm-system nanonetwork is introduced to each sample condition.
- Output Measurement: After a defined incubation period under controlled conditions (37°C, 5% CO₂ if cells are present), the network output is measured. For optical systems, this is via fluorescence plate reader or flow cytometry; for magnetic systems, via NMR relaxometry.
- Data Analysis: Dose-response curves are fitted to calculate the limit of detection (LOD), dynamic range, and EC₅₀.
Table 1: Exemplar Ex Vivo Validation Data for a Prototype IL-6 Sensing Nanonetwork
| Sample Matrix | Spiked [IL-6] (pg/mL) | Mean Net RFU* | SD | % Recovery |
|---|---|---|---|---|
| Healthy Serum | 0 | 150 | 12 | N/A |
| Healthy Serum | 10 | 520 | 45 | 98% |
| Healthy Serum | 100 | 3200 | 210 | 102% |
| Healthy Serum | 1000 | 12500 | 980 | 101% |
| ARDS Serum | (Endogenous) | 4150 | 320 | (Quantified) |
| Calculated Metrics | LOD: 2.5 pg/mL | Dynamic Range: 5-5000 pg/mL | EC₅₀: 85 pg/mL |
RFU: Relative Fluorescence Units; *ARDS: Acute Respiratory Distress Syndrome*
Animal Model Validation: Correlating Output with Disease ProgressionIn Vivo
Core Experimental Protocol
- Objective: To demonstrate that temporal changes in nanonetwork output correlate with disease onset, progression, or response to therapy in a living organism.
- Methodology:
- Model Selection: Employ a relevant animal model (e.g., murine colitis model, orthotopic tumor xenograft, LPS-induced inflammation).
- Nanonetwork Administration: The nanonetwork is delivered via a relevant route (e.g., intravenous, intraperitoneal).
- Longitudinal Output Monitoring: At predetermined timepoints, the network output is measured non-invasively (e.g., fluorescence molecular tomography, whole-body imaging) or via terminal sampling of blood/tissues.
- Endpoint Correlative Histopathology: Animals are euthanized; target tissues are harvested for gold-standard analysis (IHC, ELISA, RNA-seq) to establish a direct spatial and quantitative correlation with the in vivo nanonetwork signal.
Table 2: Longitudinal Data from a Murine Tumor Apoptosis Model
| Day Post-Treatment | Treatment Group | Mean Nano-Signal (p/s/cm²/sr) | SD | Ex Vivo Tumor Caspase-3 (pmol/min/µg) | Tumor Volume (mm³) |
|---|---|---|---|---|---|
| 0 (Baseline) | Control | 1.2e⁵ | 1.1e⁴ | 0.15 | 150 |
| 0 (Baseline) | Therapeutic | 1.3e⁵ | 1.5e⁴ | 0.18 | 155 |
| 2 | Control | 1.4e⁵ | 1.3e⁴ | 0.21 | 320 |
| 2 | Therapeutic | 8.9e⁶ | 7.8e⁵ | 2.85 | 290 |
| 5 | Control | 1.8e⁵ | 1.6e⁴ | 0.25 | 610 |
| 5 | Therapeutic | 3.2e⁶ | 2.9e⁵ | 1.20 | 220 |
Pearson correlation between Nano-Signal and Caspase-3 activity: r = 0.94 (p < 0.001).
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Validation Studies
| Item | Function/Application | Example (Supplier) |
|---|---|---|
| Recombinant Biomarker Proteins | For spiking experiments and standard curve generation in ex vivo assays. | Human IL-6, TNF-α (R&D Systems) |
| Disease-Specific Animal Models | Provide a pathophysiologically relevant in vivo environment for testing. | IL-10⁻/⁻ Colitis Mouse Model (The Jackson Laboratory) |
| Validated ELISA or MSD Assay Kits | Gold-standard method to confirm biomarker levels in ex vivo samples and tissue lysates. | V-PLEX Proinflammatory Panel 1 (Meso Scale Discovery) |
| Multispectral Imaging System | Enables longitudinal, quantitative measurement of optical nanonetwork output in vivo. | IVIS Spectrum (PerkinElmer) |
| Flow Cytometry Antibody Panels | For characterizing nanonetwork-cell interactions and immune context in harvested tissues. | Anti-mouse CD45, CD11b, F4/80 (BioLegend) |
| Tissue Digestion & Homogenization Kits | Prepare tissue samples for downstream correlative biomarker analysis. | GentleMACS Dissociator (Miltenyi Biotec) |
Visualizing the Validation Workflow & Signaling
Validation Cascade from Bench to In Vivo
Title: Sequential Validation Workflow for Diagnostic Nanonetworks
Key Signaling Pathway Correlated in Inflammatory Model
Title: TLR4-NF-κB Pathway Linked to Nanonetwork Output
Within the thesis framework of a basic alarm-system nanonetwork for biomarker research, two dominant paradigms for molecular-scale communication and computation have emerged: DNA-based nanocommunication and synthetic enzyme-powered networks. These systems are engineered to detect specific biomarkers (e.g., mRNA, proteins, metabolites) in situ and propagate a detectable signal, forming the core of a diagnostic or research nanomachine. This whitepaper provides a technical comparison of their core architectures, experimental protocols, and implementation toolkits.
Core Architectures & Signaling Pathways
DNA Nanocommunication Networks
DNA networks use engineered nucleic acid strands as information carriers and logic gates. Communication occurs via strand displacement reactions, where an input DNA strand binds to a gate complex, displacing and releasing an output strand that serves as the signal for the next node. A typical alarm system cascade involves a biomarker-triggered initiator strand that propagates through a series of logic gates, culminating in the amplified release of a reporter strand (e.g., for fluorescence readout).
Diagram Title: DNA Nanocommunication Alarm Cascade
Synthetic Enzyme-Powered Networks
These networks employ synthetic or engineered enzymes (e.g., DNAzymes, ribozymes, or allosteric protein enzymes) as signal processors. A biomarker binding event allosterically activates an enzyme, which then catalyzes the conversion of a substrate into a product. This product can act as a diffusible messenger (e.g., a small molecule) to activate downstream enzyme nodes, creating a communication cascade. Signal amplification is intrinsic to the catalytic turnover.
Diagram Title: Enzyme-Powered Network Signaling
Quantitative Comparison Table
| Parameter | DNA Nanocommunication Networks | Synthetic Enzyme-Powered Networks |
|---|---|---|
| Primary Signal Carrier | Nucleic Acid Strands (Logic Gates) | Small Molecules / Ions |
| Communication Speed | ~10^-3 to 10^-1 s^-1 (for strand displacement) | ~10^2 to 10^5 s^-1 (catalytic turnover) |
| Signal Amplification Mechanism | Autonomous hybridization chain reaction (HCR) or CRISPR | Intrinsic enzyme catalysis |
| Effective Range | Short-range (diffusion-limited, nanometers to microns) | Medium-range (diffusible messengers, up to ~100 µm) |
| Power Source | Chemical potential of annealed strands | Chemical energy of substrate cleavage/phosphorylation |
| Background Noise | Low (high specificity Watson-Crick pairing) | Moderate (potential for off-target catalysis) |
| Modularity & Design Toolkits | High (nucleic acid sequence design software, e.g., NUPACK) | Moderate (requires protein/DNAzyme engineering) |
| Typical Output Signal | Fluorescent, FRET-based, or electrochemical | Colorimetric, fluorescent, or chemiluminescent |
| Stability in Complex Biofluids | Moderate (susceptible to nucleases) | Variable (protein enzymes susceptible to proteolysis) |
Experimental Protocols for Core Function Validation
Protocol: Validating a DNA Strand Displacement Alarm Cascade
Objective: To confirm biomarker-mimic initiator strand triggers a full cascade resulting in fluorescent output.
- Materials: See "Scientist's Toolkit" below.
- Preparation: Resuspend all DNA strands (gate, inhibitor, reporter, fuel) in nuclease-free TE buffer. Anneal gate complexes by heating to 95°C for 5 min and slowly cooling to 25°C over 90 min.
- Reaction Setup: In a black 384-well plate, combine:
- 50 nM annealed gate complex
- 100 nM fluorescent reporter strand (quenched)
- 500 nM fuel strands (in excess)
- 1x reaction buffer (20 mM Tris-HCl, 150 mM NaCl, 5 mM MgCl2, pH 7.5).
- Initiation: Add the target initiator strand (biomarker mimic) at concentrations from 0 to 100 nM. Run negative control (no initiator).
- Data Acquisition: Monitor real-time fluorescence (e.g., FAM channel) in a plate reader at 37°C for 2-4 hours.
- Analysis: Calculate reaction initiation time and final fluorescence fold-change over background.
Protocol: Validating an Enzyme-Powered Network Cascade
Objective: To confirm allosteric activation of a DNAzyme by a target and subsequent substrate turnover.
- Materials: See "Scientist's Toolkit" below.
- DNAzyme Preparation: Anneal the allosteric DNAzyme strand with its substrate arm.
- Reaction Setup: In a clear 96-well plate, combine:
- 100 nM annealed DNAzyme complex
- 1 µM caged fluorogenic substrate (e.g., RNA-linked fluorescein)
- 1x reaction buffer (50 mM HEPES, 150 mM NaCl, 20 mM MgCl2, pH 7.0).
- Initiation: Add the target biomarker (or mimic) at varying concentrations.
- Data Acquisition: Immediately measure fluorescence (Ex/Em ~490/520 nm) every 30 seconds for 60 minutes at 37°C.
- Analysis: Determine initial reaction velocity (V0) and plot against target concentration to derive activation kinetics.
The Scientist's Toolkit: Essential Research Reagents
| Item Name / Category | Function in Experiments | Example Vendor(s) |
|---|---|---|
| DNA Oligonucleotides | Synthesized strands for gates, initiators, and reporters; backbone of DNA networks. | IDT, Sigma-Aldrich |
| Fluorophore-Quencher Pairs | For constructing molecular beacons and quenched reporter substrates (e.g., FAM/BHQ-1). | Lumiprobe, Biosearch Tech |
| Nuclease-Free Buffers & Water | Essential for preparing and diluting nucleic acid components to prevent degradation. | Thermo Fisher, Ambion |
| Custom DNAzymes/Ribozymes | Engineered catalytic nucleic acids for synthetic enzyme networks. | Bio-Synthesis Inc. |
| Caged Fluorogenic Substrates | Silent probes that yield fluorescence upon enzymatic cleavage (e.g., RNA-FAM). | AAT Bioquest, Cayman Chem |
| Real-Time PCR/Plate Reader | Instrumentation for kinetic measurement of fluorescent output signals. | Bio-Rad, Agilent |
| CRISPR-Cas Components (e.g., Cas12a/13a) | For high-gain amplification modules in advanced DNA networks. | New England Biolabs |
| Microfluidic Chips | For testing network function in confined geometries mimicking physiological environments. | Dolomite, Fluigent |
Within the broader thesis on the Basic Architecture of an Alarm-System Nanonetwork for Biomarkers Research, a critical evaluation of established analytical techniques is essential. This nanonetwork aims to provide real-time, in vivo surveillance of biomarkers, a paradigm shift from traditional ex vivo or endpoint assays. To contextualize this shift, we present a comparative technical analysis of three cornerstone methodologies: Enzyme-Linked Immunosorbent Assay (ELISA), Polymerase Chain Reaction (PCR), and Imaging (focusing on fluorescence and bioluminescence). This guide examines their principles, protocols, and quantitative performance metrics, highlighting the operational gaps that advanced nanonetworks are designed to address.
Core Principles and Comparative Metrics
Fundamental Assay Architectures
ELISA operates on the principle of antigen-antibody binding, with an enzyme-mediated colorimetric, chemiluminescent, or fluorescent readout. It is the gold standard for protein quantification. PCR amplifies specific DNA sequences exponentially via thermal cycling, enabling the detection of nucleic acid biomarkers with extreme sensitivity. Imaging (Bioluminescence/Fluorescence) utilizes light-emitting reporters (luciferases, fluorescent proteins/dyes) to visualize spatial and temporal biomarker distribution in vivo.
Quantitative Performance Comparison
Table 1: Comparative Performance Metrics of Traditional Assays
| Parameter | ELISA | qPCR/dPCR | In Vivo Imaging |
|---|---|---|---|
| Target Type | Proteins, peptides, antibodies | DNA, RNA, methylated DNA | Cells, enzymes, gene expression |
| Sensitivity | ~pg/mL (10⁻¹² g/mL) | aM- fM (10⁻¹⁸ - 10⁻¹⁵ M) for dPCR | ~10³ - 10⁴ cells (luminescence) |
| Dynamic Range | 2-3 logs | 7-8 logs (qPCR) | 3-4 logs |
| Multiplexing Capacity | Low-Moderate (up to ~10-plex) | Moderate-High (up to 50-plex with ddPCR) | Low (2-3 colors in vivo) |
| Temporal Resolution | Endpoint (single time point) | Endpoint (single time point) | Real-time (longitudinal) |
| Spatial Resolution | None (homogenized sample) | None (homogenized sample) | Millimeters (whole-body) to microns (IVIS/CT) |
| Throughput | High (96/384-well plates) | High (96/384-well plates) | Low (sequential animal imaging) |
| Key Advantage | Specific, quantitative, high-throughput | Ultra-sensitive, specific, quantitative | Non-invasive, longitudinal, provides context |
| Key Limitation | No spatial/temporal data, requires lysis | No spatial/temporal data, requires lysis | Limited sensitivity and depth penetration |
Detailed Experimental Protocols
Protocol: Sandwich ELISA for Cytokine Detection
- Coating: Coat a 96-well plate with 100 µL/well of capture antibody (1-10 µg/mL in carbonate buffer). Incubate overnight at 4°C.
- Blocking: Aspirate, wash 3x with PBS + 0.05% Tween-20 (PBST). Add 200 µL blocking buffer (e.g., 5% BSA in PBS). Incubate 1-2 hours at RT.
- Sample Incubation: Wash 3x. Add 100 µL of sample or standard (serially diluted in dilution buffer). Incubate 2 hours at RT or overnight at 4°C.
- Detection Antibody Incubation: Wash 3x. Add 100 µL of biotin-conjugated detection antibody. Incubate 1-2 hours at RT.
- Streptavidin-Enzyme Conjugate: Wash 3x. Add 100 µL of Streptavidin-HRP (1:5000 dilution). Incubate 30 minutes at RT in the dark.
- Signal Development: Wash 3x. Add 100 µL TMB substrate. Incubate 10-20 minutes at RT.
- Stop & Read: Add 50 µL 2N H₂SO₄ to stop reaction. Measure absorbance immediately at 450 nm with a reference at 570 nm.
Protocol: Reverse Transcription Quantitative PCR (RT-qPCR)
- RNA Isolation: Lyse cells/tissue in TRIzol. Perform chloroform phase separation. Precipitate RNA with isopropanol, wash with 75% ethanol, and resuspend in RNase-free water. Quantify via spectrophotometry.
- DNase Treatment: Treat 1 µg RNA with DNase I (RNase-free) for 15 min at RT. Inactivate with EDTA and heat.
- Reverse Transcription: Use a high-capacity cDNA reverse transcription kit. Mix RNA, random hexamers, dNTPs, buffer, and reverse transcriptase. Incubate: 25°C (10 min), 37°C (120 min), 85°C (5 min).
- qPCR Setup: Prepare a master mix containing SYBR Green PCR Master Mix, forward/reverse primers (200-500 nM final), nuclease-free water, and cDNA template (~10-100 ng equivalent).
- Thermal Cycling: Run in a real-time PCR system: Initial denaturation: 95°C, 10 min; 40 cycles of: 95°C for 15 sec (denature), 60°C for 1 min (anneal/extend). Acquire SYBR Green signal at the end of each extension step.
- Analysis: Determine cycle threshold (Ct) values. Use a standard curve or ΔΔCt method for relative quantification.
Protocol: In Vivo Bioluminescence Imaging (BLI) of Tumor Cells
- Reporter Engineering: Stably transduce target cells (e.g., cancer cells) with a lentivirus expressing firefly luciferase (Fluc).
- Animal Model: Inject labeled cells (e.g., 1x10⁵ in 100 µL PBS) subcutaneously or intravenously into immunodeficient mice.
- Substrate Administration: Anesthetize mouse with isoflurane. Inject D-luciferin substrate intraperitoneally (150 mg/kg in PBS).
- Imaging: Place mouse in the imaging chamber (IVIS Spectrum or equivalent) 10-15 minutes post-injection. Acquire a grayscale photograph followed by a bioluminescent image (exposure: 1 sec to 5 min, depending on signal).
- Image Analysis: Use Living Image or similar software. Define regions of interest (ROI) over signal areas. Quantify total flux (photons/sec).
Signaling Pathways and Workflows
Diagram Title: Comparative Workflows for ELISA, PCR, and Imaging
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagent Solutions for Featured Assays
| Reagent/Material | Primary Assay | Function/Brief Explanation |
|---|---|---|
| High-Affinity Capture & Detection Antibodies | ELISA | Provide specificity for the target antigen. Critical for sensitivity and low background. |
| Recombinant Protein Standards | ELISA | Quantification reference. Must be highly pure and accurately quantified. |
| HRP-Conjugated Streptavidin & TMB Substrate | ELISA | Signal generation system. Streptavidin binds biotin; HRP catalyzes colorimetric TMB reaction. |
| TRIzol / Guanidine Thiocyanate Lysis Buffer | PCR | Simultaneously denatures proteins and protects RNA from RNases during cell lysis. |
| DNase I (RNase-free) | PCR | Removes genomic DNA contamination from RNA preparations to prevent false-positive PCR signals. |
| SYBR Green Master Mix | qPCR | Contains Hot Start Taq, dNTPs, buffer, and SYBR Green dye. Binds double-stranded DNA for detection. |
| D-Luciferin, Potassium Salt | BLI | Cell-permeable substrate for firefly luciferase. Reaction with luciferase/O₂ produces light. |
| Isoflurane / Anesthetic System | BLI | Maintains animal sedation and immobility during image acquisition for consistent results. |
| IVIS Spectrum or Equivalent Imager | BLI | Cooled CCD camera system capable of detecting low-intensity bioluminescent and fluorescent light from live animals. |
ELISA, PCR, and Imaging each offer powerful, yet fundamentally limited, windows into biomarker biology. ELISA and PCR provide exceptional sensitivity and quantification but sacrifice spatial and temporal context. Imaging offers longitudinal and spatial data but lacks the quantitative rigor and multiplexing depth of the other techniques. The proposed alarm-system nanonetwork architecture seeks to synthesize the strengths of these modalities: the specificity of immunoassays, the amplification potential of molecular circuits, and the real-time, in situ reporting of imaging, thereby enabling a transformative continuous surveillance platform for biomarker research and therapeutic intervention.
Evaluating Clinical Translational Potential and Regulatory Pathways
Within the architectural framework of an alarm-system nanonetwork for biomarkers research, the ultimate measure of success is its translation into clinical practice. This nanonetwork, designed for in vivo detection, amplification, and reporting of specific biomarker signatures, represents a disruptive diagnostic paradigm. However, its complexity necessitates a rigorous, parallel evaluation of both its clinical translational potential and the regulatory pathways required for approval. This guide provides a structured, technical approach for researchers to navigate this critical stage, transforming a proof-of-concept into a viable Investigational Device.
Quantitative Framework for Translational Potential Assessment
The clinical value of an alarm-system nanonetwork must be quantitatively de-risked across multiple dimensions. The following tables consolidate key metrics that must be empirically established.
Table 1: Analytical and Preclinical Performance Benchmarks
| Metric | Target Specification | Experimental Method | Relevance to Translation |
|---|---|---|---|
| Limit of Detection (LoD) | ≤ 1 pM for target biomarker | Dose-response in simulated matrix (PBS, serum) | Determines earliest disease stage detectable. |
| Dynamic Range | ≥ 4 orders of magnitude | Dose-response curve analysis | Ensures quantification across clinically relevant concentrations. |
| Signal-to-Background Ratio | > 10:1 in vivo | Comparison in target vs. control tissue (animal model) | Critical for reliable alarm triggering. |
| Nanosensor Pharmacokinetics | Circulation t½ > 30 min; Clearance < 24h | Radiolabeling & bio-distribution study (IVIS, SPECT) | Informs dosing and safety. |
| Immunogenicity Risk | Negligible anti-nanoparticle antibody response | ELISA for IgG/IgM post-administration (animal) | Major safety and efficacy concern. |
| Target Selectivity | >100-fold vs. closest homolog | Cross-reactivity panel assay | Prevents false-positive alarms. |
Table 2: Preliminary Clinical Utility & Commercial Viability Assessment
| Criterion | Key Questions | Data Sources |
|---|---|---|
| Unmet Clinical Need | Does it enable earlier intervention or guide therapy where current diagnostics fail? | Clinical guidelines, key opinion leader (KOL) interviews. |
| Intended Use & Claim | Can a clear, specific diagnostic claim be defined? (e.g., "detects micrometastases >3mm") | Regulatory precedent (FDA Decision Summaries). |
| Target Population | Size, accessibility, and standard of care. | Epidemiological databases, market reports. |
| Health Economic Value | Will it reduce overall cost of care or enable cost-effective screening? | Cost-effectiveness model (CEA) draft. |
| Reimbursement Likelihood | Is there an existing CPT code, or will a new one be needed? | Payer policy reviews (CMS, private). |
Experimental Protocols for Critical Translational Studies
Protocol 1: In Vivo Specificity and Off-Target Activation Assessment
- Objective: To quantify the nanonetwork's activation in non-target tissues and in the presence of confounding physiological states (e.g., inflammation).
- Materials: Animal disease model, control (healthy) animals, fluorescent or bioluminescent reporter nanonetwork, IVIS imaging system, tissue homogenization kit, qPCR reagents (if reporter is genetic).
- Method:
- Administer nanonetwork intravenously to disease model (n=8) and healthy controls (n=8).
- Allow for circulation and biomarker interaction (time determined by PK studies).
- Perform whole-body longitudinal imaging at 0, 6, 12, 24h post-injection.
- Euthanize animals, harvest target and non-target organs (liver, spleen, kidney, lung).
- Image ex vivo organs and quantify signal intensity (photons/sec/cm²/sr).
- Homogenize tissues and quantify reporter signal (luminescence, fluorescence, or mRNA) biochemically for normalization.
- Statistically compare target vs. non-target signal in disease models, and target signal in disease vs. healthy controls.
Protocol 2: Dose-Ranging and Minimum Effective Dose (MED) Study
- Objective: To establish the relationship between nanonetwork dose and output signal magnitude, defining the MED for reliable alarm generation.
- Materials: Animal disease model, nanonetwork stock at high concentration, sterile PBS for dilution, imaging system.
- Method:
- Prepare 4-5 log-spaced doses of the nanonetwork (e.g., 10¹⁰, 10¹¹, 10¹² particles/kg).
- Randomly assign animals (n=5 per dose group) to receive one dose via IV.
- Image at the predetermined peak signal time.
- Quantify the target site signal and plot against administered dose.
- Fit a sigmoidal dose-response curve. The MED is defined as the dose producing a signal significantly above background (p<0.05) and at 90% of the maximal response (EC90).
Mapping the Regulatory Pathway: A Dual-Track Strategy
For an alarm-system nanonetwork, regulatory strategy is not an afterthought but a core component of the experimental design. The pathway is typically determined by its risk classification and intended use.
Diagram 1: Core US FDA Regulatory Decision Pathway for a Diagnostic Nanonetwork
Key Regulatory Experiments:
- Analytical Validation: Conduct per CLSI guidelines (EP05, EP06, EP07, EP17) to generate data for a 510(k) or Premarket Approval (PMA) submission.
- Bench Top Failure Mode Analysis: Document all potential failure modes of the nanonetwork logic (e.g., non-specific activation, premature release).
- Stability & Shelf-Life Studies: Real-time and accelerated stability testing of the nanonetwork formulation under defined storage conditions.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Translational Nanonetwork Research
| Item | Function & Rationale |
|---|---|
| PEGylated Lipid Nanoparticles | The foundational delivery vehicle. PEGylation ("stealth" coating) extends circulation half-life and reduces immune clearance, critical for in vivo efficacy. |
| Activation-Specific Linker Chemistry | The "alarm trigger." Cleavable linkers (e.g., protease-sensitive, pH-sensitive) connecting reporter to nanoparticle core must have high specificity for the target biomarker. |
| Near-IR Fluorophores / Luciferin-Luciferase | Reporters for deep-tissue imaging. Near-IR light penetrates tissue better. Bioluminescence (luciferase) offers extremely low background but requires substrate delivery. |
| Quantum Dots with Bioconjugation Handles | Bright, photostable alternative to fluorophores. Allow multiplexing if different biomarkers are tagged with different QD emission spectra. |
| Animal Disease Models (Orthotopic/GEMM) | Critical. Models must faithfully recapitulate human disease biology and biomarker expression patterns for translational relevance. |
| IVIS Spectrum or MSFX Imaging System | Enables longitudinal, quantitative 2D/3D imaging of fluorescence/bioluminescence in vivo in live animals. |
| Simulated Biological Matrices | (e.g., synthetic serum, interstitial fluid) Used for robust in vitro analytical validation under controlled, reproducible conditions. |
| Human Tissue Microarrays (TMAs) | Enable high-throughput validation of biomarker presence and correlation with disease stage on actual human clinical samples. |
| Anti-PEG Antibody ELISA Kit | To assess immunogenicity potential, a major risk for repeat-administration diagnostics or future therapeutic versions. |
| Size Exclusion Chromatography with MALS | For precise, quantitative characterization of nanonetwork size, aggregation state, and stability in solution. |
Diagram 2: Functional Workflow of an Alarm-System Diagnostic Nanonetwork
Conclusion
The architecture of alarm-system nanonetworks represents a paradigm shift from passive biomarker measurement to active, distributed sensing and communication. By mastering the foundational principles, methodological assembly, and rigorous optimization outlined here, researchers can transition from proof-of-concept to robust, deployable systems. While challenges in signal fidelity, biocompatibility, and *in vivo* validation remain, the convergence of nanotechnology, synthetic biology, and information theory is rapidly providing solutions. The future lies in intelligent, closed-loop systems where nanonetworks not only detect but also initiate therapeutic responses. For drug developers, these networks offer unprecedented tools for real-time pharmacokinetic/pharmacodynamic monitoring in clinical trials, paving the way for a new era of precision medicine and proactive healthcare.